
Laboratory of protein spectroscopy
The physical basis of protein function
The physical factors behind protein function are the electrostatic interaction between charges and dipoles; electron affinity and the related electron transfer reactions; proton affinity (pK) and the related proton transfer reactions as well as hydrogen bonds; steric interactions depending on the 3D protein structure; as well as the interactions with the water molecules inside and around the protein and with the surrounding lipid molecules in case of the membrane proteins. For proteins with chromophoric prostetic groups spectroscopic and kinetic spectroscopic studies in the visible-UV range provide essential information about the protein function.
We study the photocycle and the molecular steps of the proton transport in bacterial retinal protein proton pumps, e.g. bacteriorhodopsin, or in signaling proteins, e.g. PYP, and their dependence on environmental factors. We investigate the structural factors affecting the kinetics of electron transfer in cytochromes. We develop and apply general chemometric methods for the analysis of spectral data matrices in order to determine the underlying physical phenomena. In addition, we study the possibilities of the integration of „colored” proteins in biophotonic and in bioelectronic applications.
Our main experimental setup consists of a tunable Nd-Yag + OPO pulse laser to provide < 10 ns actinic pulses and a continuous white measuring light source in the entire visible range. Time resolved spectra of the excited sample can be measured at delay times from <100ns upwards. High time resolution single wavelength kinetics can also be measured in the same experimental arrangement.
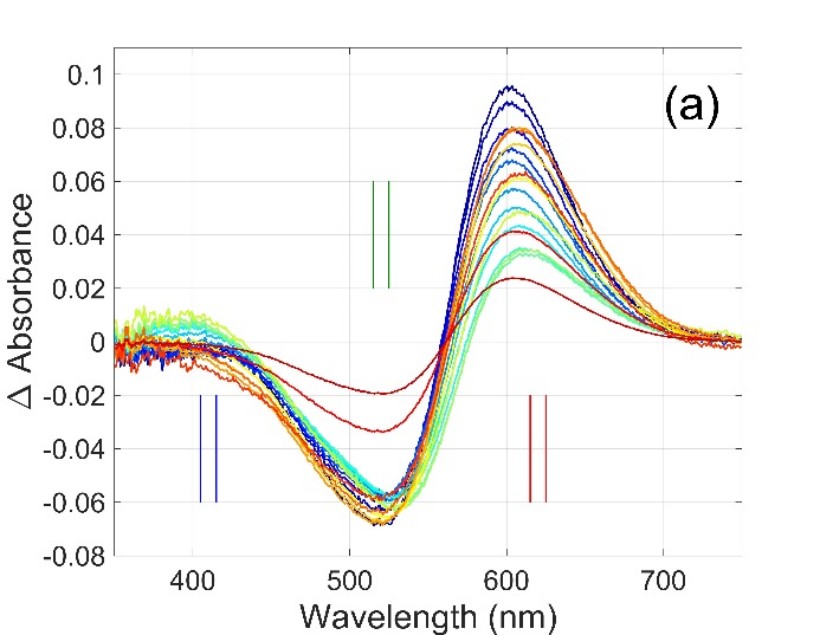
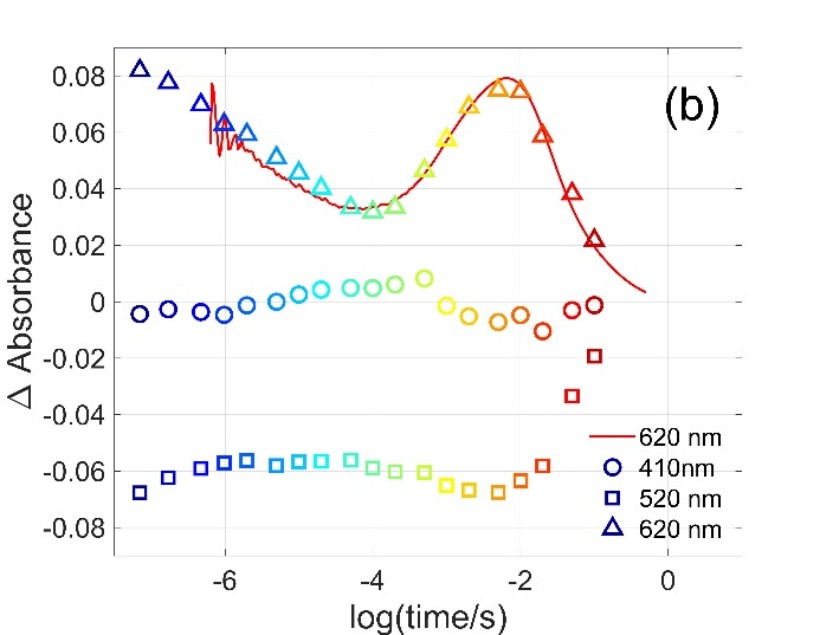
Figure 1. Multiwavelength and single wavelength absorption change signals measured on Gloeobacter rhodopsin [1]. Symbols in (b) are the amplitudes obtained at the selected wavelengths from the difference spectra in (a).
Structure-function relationships of cytochrome b561 proteins
Cytochrome b561 proteins are ascorbate reducible, 6 transmembrane helix proteins in eukaryotes housing 2 heme b centers close to the opposite membrane surfaces. They are reduced on the cytoplasmic side by ascorbate, and after transporting the electron via the hemes to the non-cytoplasmic side, reduce monodehydroascorbate, ferric chelates or so far unidentified electron acceptors. We have measured spectral and redox properties as well as substrate titration parameters of several members of this protein family, and we study the role of conserved amino acid residues and structural features relevant to their function based on homology modeling and AlphaFold structures.
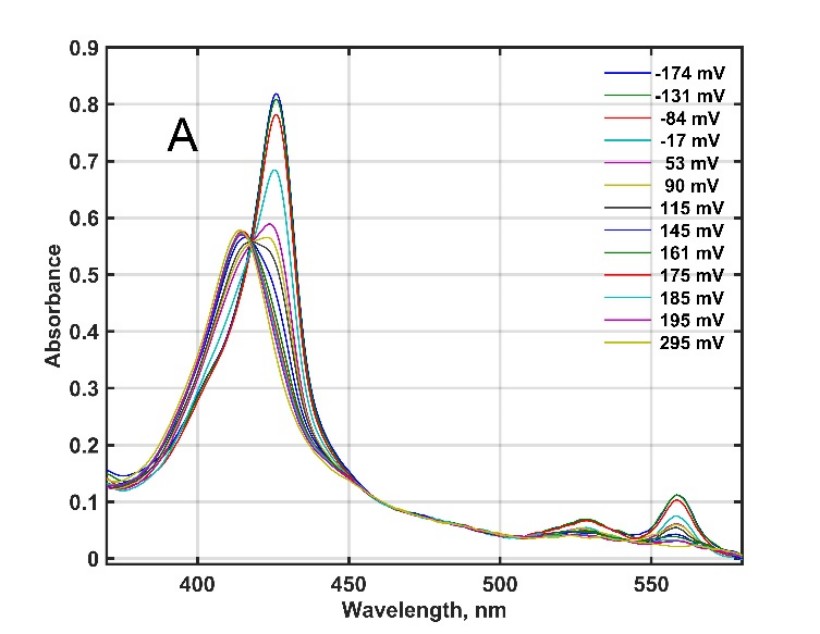
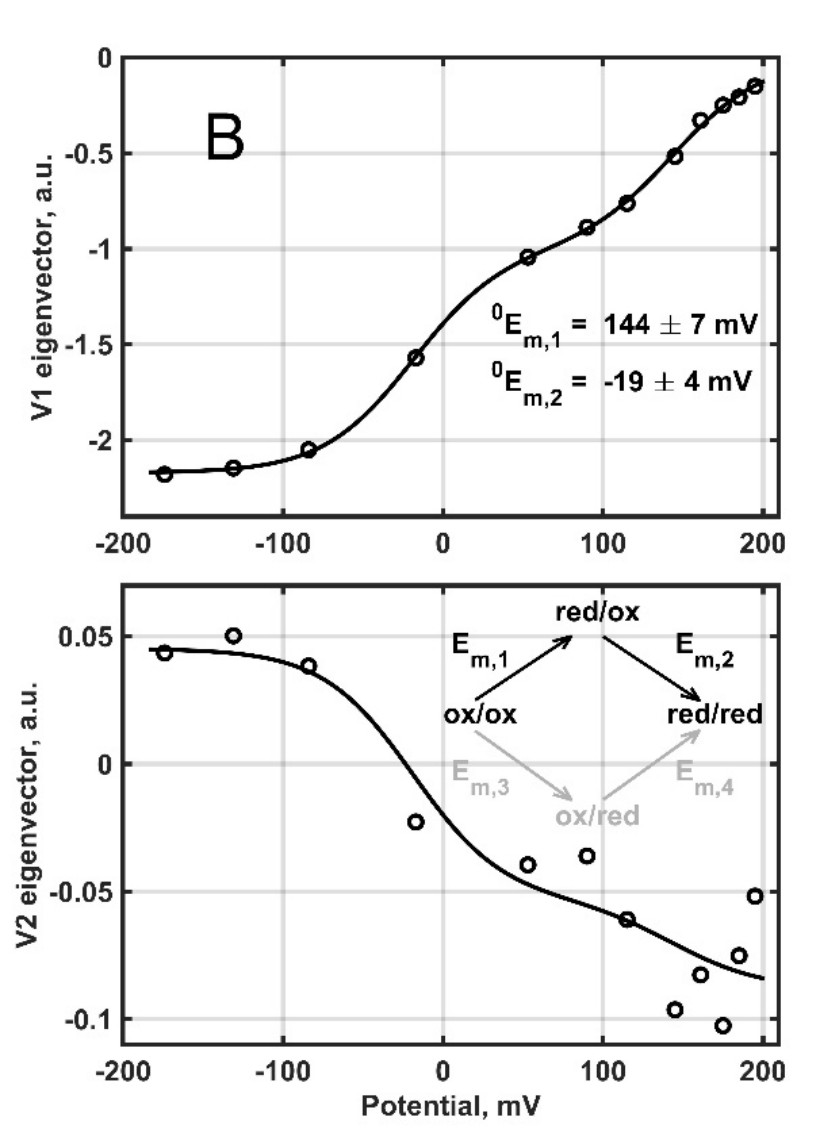
Figure 2. Redox titration and a general model of successive reduction of mouse cytochrome b561 D1 with so far unknown physiological function [2].
Porous silicon based biophotonics
We successfully created a composite biophotonic structure out of porous silicon (PSi) microcavities doped by the photochromic protein, photoactive yellow protein (PYP) [3]. With this proof-of-concept design we could demonstrate light induced optical response (reflectance change) in the microsecond range, consistent with the nonlinear optical properties and the photocycle kinetics of the incorporated protein.
Femtobiology laboratory
Fluorescence spectroscopy with femtosecond time resolution
Recently we constructed a measuring system suitable for the detection of fluorescence kinetics in the 100 fs – 10 ns time interval and in the complete visible spectral range. On this apparatus presently we are focusing on the study of the coenzymes FAD and NADH – substantial components of redox systems – in different microenvironments. We have found that in aqueous solution the kinetics of FAD fluorescence (Fig. 3) can be characterized by several time constants: one of them, falling in the ns range, can be associated to the open conformation of the molecule, while three components in the ps range correspond to closed conformations. The Hofmeister effect – controlling the ratio of molecules in the different conformational states – is well observable on the kinetics, in accordance with the models based on molecular dynamics calculations. The setup also enables the observation of the Stokes-shift, characterized by a lifetime of ~100 fs, and the study of the related solvation dynamics. In contrast to the above parameters, in the case of FAD bound covalently to the flavocytochrome c sulfide dehydrogenase enzyme, we observed only a single sub-ps component, indicating that the coenzyme is in an extremely strong interaction with the protein microenvironment. For coenzyme NADH the equilibrium of the open and closed conformations is also traceable, however, the Stokes-shift has three components, related to an unusually complex process of vibrational relaxation.
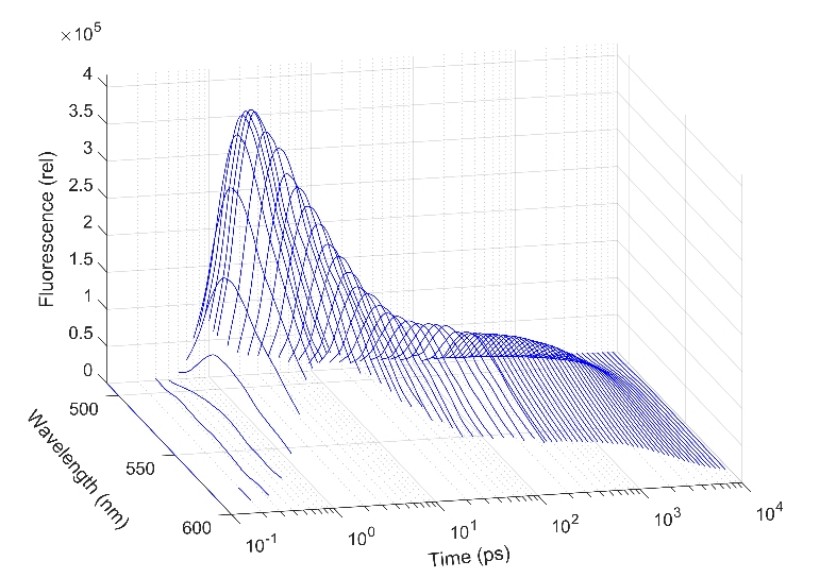
Figure 3. Fluorescence kinetics of FAD in time and wavelength domain
Analysis of complex first-order reaction kinetics systems
The above-mentioned reactions, taking place in the excited state, can be characterized by first-order kinetics, similarly to the light-induced ground-state reactions of retinal proteins. The standard way of the analysis of such complex system reactions is based on the method of nonlinear regression with exponential terms, whose application faces many difficulties. The major problem is that the solution of the fitting task is an ill-posed problem: small noise on the experimental data generates high uncertainties in the predicted parameters, and even in the number of the necessary parameters. Applying simulated data based on a very complex model of the bacteriorhodopsin photocycle we elaborated a new method for handling this problem [5]. The essential part of this procedure is that instead of discrete exponential components, the solution is characterized by a distribution over the continuous space of time-constants, and the instability of the regression is handled by adding two regularizing terms. The weight of the first regularizing term () controls the number of peaks of the distribution, and hence the complexity of the selected model, while the second one () determines their width. Applying the techniques of modern statistics (cross-validation, Bayesian optimization) we developed a machine learning method, which can automatically determine the proper value of these hyperparameters, exclusively from the simulated experimental data and the level of noise corrupting them (Fig. 4). This method also performs very well on the analysis of the experimentally detected fluorescence kinetics.
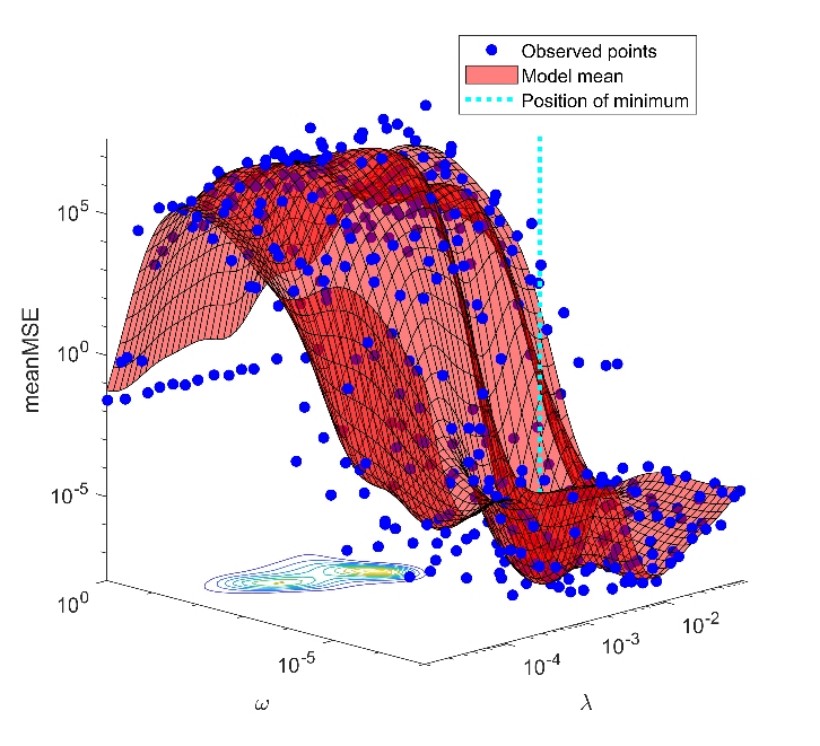
Figure 4. Selection of the hyperparameters at the minimum of cross-validation error
Development of a multifunctional femtobiology workstation
Made possible by a grant acquired recently and based on the unique infrastructure of the ELI ALPS Research Institute in Szeged, collaborating with the groups of BRC and the University of Pécs we are in the process of developing a permanent workstation with the purpose of studying ultrafast processes on biological systems utilizing light energy (light-harvesting complexes and photosynthetic reaction centers, retinal proteins, plant photoreceptors). The measuring techniques to be applied are founded partially on the expansion of the most modern technology of femtosecond multidimensional electronic spectroscopy, and partially on the improvement of the methods we developed for the detection of low-intensity light-induced coherent THz radiation. On concluding with the complete implementation, the workstation will be publicly available for users.
AFM and Raman microscopy laboratory
Biological applications of Scanning Probe and Confocal Raman Microscopy
Pioneering work was done by Dr. György Váró to establish the scanning probe laboratory within the Institute of Biophysics. Following his path, the laboratory performs structural, morphological and mechanical investigation of biological objects, ranging from single protein molecules up to living cells and tissues.
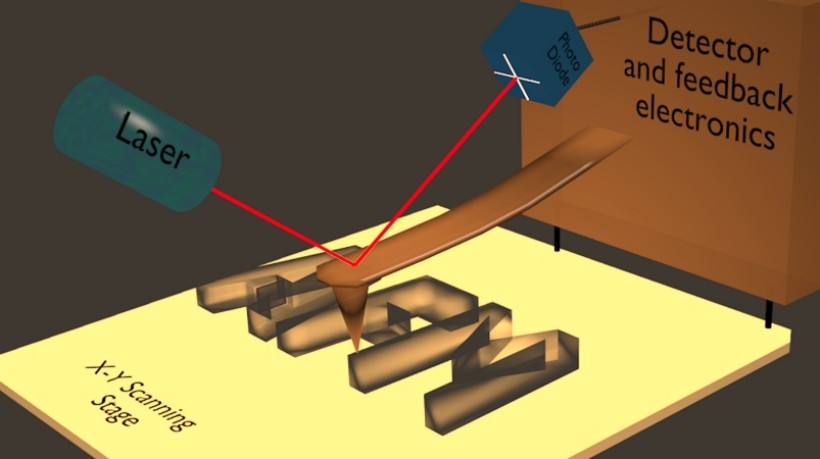
Figure 5. Schematic representation of the working principle of the Atomic Force Microscope
Nanometer scale mechanical manipulation and measurement in a liquid environment provides an absolute advantage compared to conventional cellular imaging techniques. The wide operation scale offers investigation of pico-Newton range intermolecular forces up to nano-Newton intercellular forces, even micro-Newton cell to substrate forces. Towards the understanding and the description of tumors and metastasis formation, mechanical investigations have recently gained increasing interest [8]. The ability to directly investigate the interaction of tumor cells with their environment offers outstanding opportunities. One of the active topics of the laboratory is the investigation of the mechanical interaction between tumor cells and brain endothelial cells. Figure 6 depicts a reconstructed false colored adhesion map recorded with a tumor cell as a probe.
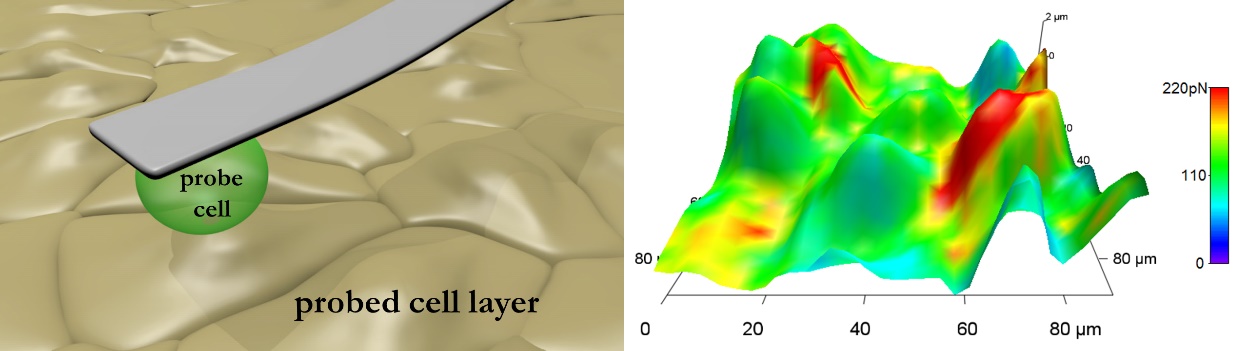
Figure 6. Left: schematic representation of the measurement setup. Right: adhesion force-colored height reconstruction of endothelial cells recorded with a living melanoma cell as a probe [9].
Atomic force microscopy augmented by label-free chemical imaging based on Raman scattering is a very potent physicochemical technique also for biological material. The method has emerged as a non-destructive and non-invasive tool to intracellularly detect administered drugs without exogenous labelling. Among the most promising applications one can find the ability of discrimination between normal and tumor cells not only in vitro, but in vivo as well. Co-localized topography, stiffness and chemical maps offer outstanding opportunities to investigate structural and dynamic processes. Successful projects included self-assembly of oligonucleotides, structural and morphological studies of nano an microvesicles, mechanical investigation of different bacterial strains, plasticity of red blood cells, de-adhesion patterns of living brain endothelial cells and, finally, quantitative mapping of several tissues. The above list is far from being complete, it only serves the presentation of the large scale and subject areas where our system can provide valuable information.
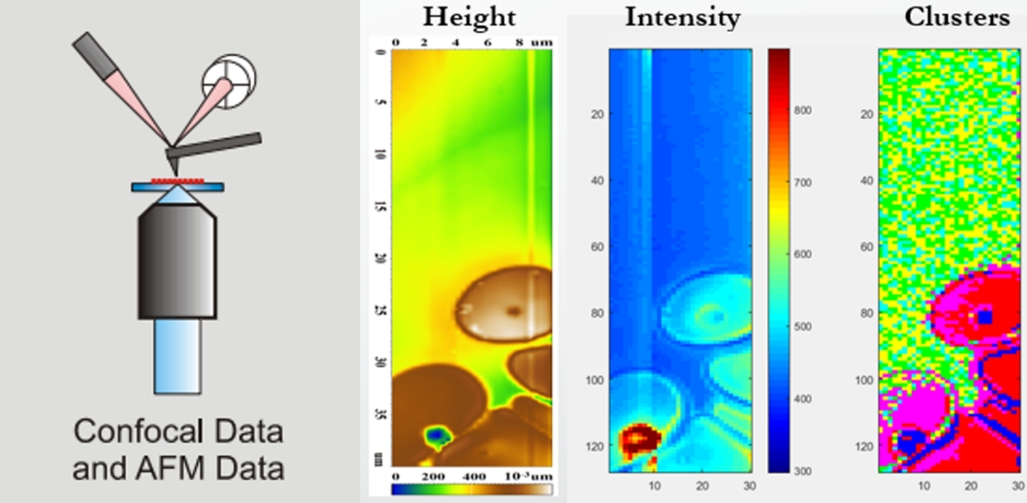
Figure 7. Scheme of the device combining confocal Raman spectroscopy and atomic force microscopy. Combined topography, Raman scattering intensity and spectral discriminative mapping of red blood cells.
Misra, R., Das, I., Dér, A., Steinbach, G., Shim, J-G., Busse, W., Jung, K-H., Zimányi, L. and Sheves, M. 2023. Impact of protein-chromophore interaction on the retinal excited state and photocycle of Gloeobacter rhodopsin: role of conserved tryptophan residues. Chemical Science 14:9951.
Bérczi, A., Márton, Z., Laskay, K., Tóth, A., Rákhely, G., Duzs, Á., Sebők-Nagy, K., Páli, T. and Zimányi, L. 2023. Spectral and redox properties of a recombinant mouse cytochrome b561 protein suggest transmembrane electron transfer function. Molecules 28(5):2261.
Petrovszki, D., Valkai, S., Kelemen, L., Nagy, L., Agarwal, V., Krekic, Sz., Zimányi, L. and Dér, A. 2023. Microsecond all-optical modulation by biofunctionalized porous silicon microcavity. Nanomaterials 13:2070.
Khoroshyy, P., Tenger, K., Chertkova, R.V., Bocharova, O.V., Kirpichnikov, M.P., Borovok, N., Groma, G.I., Dolgikh, D.A., Kotlyar, A.B. and Zimányi, L. 2021. Kinetics and energetics of intramolecular electron transfer in single-point labeled TUPS-cytochrome c derivatives. Molecules 26(22):6976.
Zimányi, L., Sipos, Á., Sarlós, F., Nagypál, R. and Groma, G. 2021. Machine learning-based model selection and parameter estimation from kinetic data of complex first-order reaction systems. PLOS One 16(8): e0255675.
Krekic, S., Zakar, T., Gombos, Z., Valkai, S., Mero, M., Zimányi, L., Heiner, Z. and Dér, A. 2020. Non-linear optical investigation of microbial chromoproteins. Front. Plant Sci 11:547818.
Zimányi, L., Thekkan, S., Eckert, B., Condren, A.R., Dmitrenko,O., Kuhn, L.R., Alabugin, I.V. and Saltiel, J. 2020. Determination of the pKa values of trans-Resveratrol, a Triphenolic Stilbene, by Singular Value Decomposition. Comparison with Theory. J. Phys. Chem. A 124(31):6294-6302.
Varga, B., Fazakas, C., Molnár, J., Wilhelm, I., Domokos, R.A., Krizbai, I.A., Szegletes, Z., Váró, G., and Végh, A.G. 2017. Direct mapping of melanoma cell - endothelial cell interactions. J. Mol. Recognit. 30(6):e2603.
Fazakas, C., Kozma, M., Molnár, K., Kincses, A., Dér, A., Fejér, A., Horváth, B., Wilhelm, I., Krizbai, I.A., and Végh, A.G. 2021. Breast adenocarcinoma-derived exosomes lower first-contact de-adhesion strength of adenocarcinoma cells to brain endothelial layer. Colloids Surf. B Biointerfaces 204, 111810.
Csonti, K., Fazakas, C., Molnár, K., Wilhelm, I., Krizbai, I.A., and Végh, A.G. 2024. Breast adenocarcinoma cells adhere stronger to brain pericytes than to endothelial cells. Coll. And Surf. B, in press.

scientific adviser

research fellow

research fellow

scientific administrator

PhD student
 László ZIMÁNYI
László ZIMÁNYI
|
scientific adviser | publications | CV |
 Attila Gergely VÉGH
Attila Gergely VÉGH
|
research fellow | publications | CV |
 Áron SIPOS
Áron SIPOS
|
research fellow | publications | CV |
 Ferenc SARLÓS
Ferenc SARLÓS
|
scientific administrator | publications | CV |
 Katalin CSONTI
Katalin CSONTI
|
PhD student | publications | CV |