Research focus
Cell adhesion plays a crucial role in numerous biological processes, including tissue formation, cell communication, and cell movement. Adhesion is influenced by intercellular interactions, the interactions between cells and the extracellular matrix, and environmental factors such as mechanical forces and chemical composition. Our research group investigates the interaction between these factors in various environmental conditions, including various buffer solutions, nutrient media, and upon enzymatic digestion of selected glycocalyx components. Understanding the dynamics of adhesion is important not only for basic research but also for applications in fields such as biomedicine, tissue engineering, and biomaterial design.
The surface of cells is covered by a glycocalyx layer, composed of various sugar molecules and proteins. This layer not only provides protection to cells but also plays a key, but not fully understood role in adhesion processes. Importantly, there is no consensus in the literature on the exact role of cell surface polysaccharides and their derivatives on cell adhesion. Different enzymatic treatments can remove or modify the components of the glycocalyx, significantly impacting the behavior of the cells. We are using targeted enzymatic digestion of some of the glycocalyx components to better understand the mechanism of cellular adhesion [1].
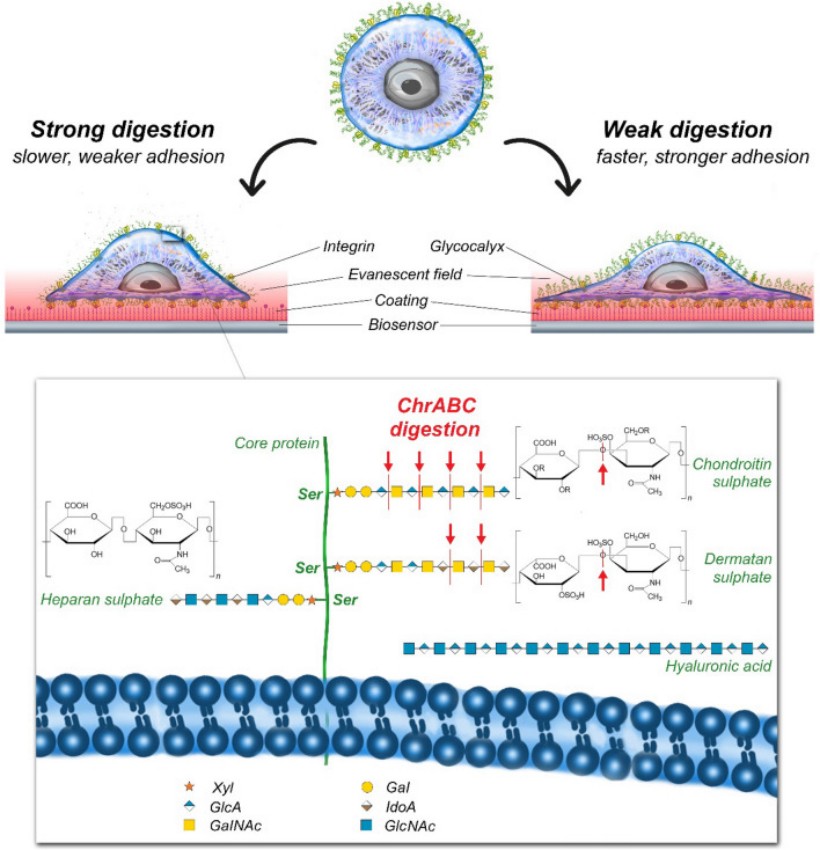
Figure 1: Enzymatic digestion of glycocalyx components and its measurement by label-free optical biosensor [1].
To study the effect of the digestion of certain glycocalyx components, the surface charge density (zeta potential) of cells is also measured. In a pilot study, we observed that the ChrABC enzyme, when applied at a certain concentration, strengthens the adhesion of cancer cells. However, at higher concentrations, the enzyme strongly weakens cell adhesion, meaning that modifying the glycocalyx layer can alter the cell's adhesion capacity in either direction, depending on the concentration of the applied enzyme [1].
This information is crucial for drug development and tissue engineering applications as it provides the opportunity to specifically influence adhesion in a novel manner.
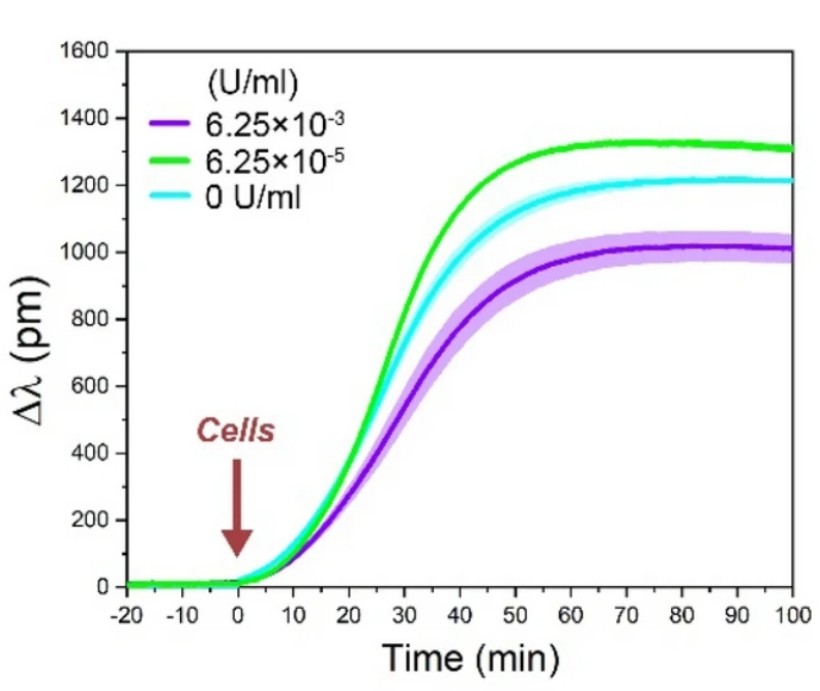
Figure 2: Representative cell adhesion kinetic curves on a 50% PP: PPR copolymer-treated surface. The cell adhesion resulted in sigmoidal shaped kinetic curves with varying magnitude, depending on the enzyme concentration: 6.15 × 10-5 U/ml enzyme increased, while 6.15 × 10-3 U/ml decreased the magnitude of the biosensor curves compared to the reference data with 0 U/ml enzyme concentration. All experiments were conducted in triplicates, and data are presented as mean ± SD [1].
Cell Adhesion in Static and Dynamic Environments
In static environments, cell adhesion processes are primarily determined by cell surface receptors, extracellular matrix proteins, and changes in the structure of the cell membrane. In such environments, cells gradually stabilize on the surface, and interactions occur under relatively constant forces. The static environment allows for the long-term establishment of adhesive contacts, making it possible to investigate the resulting morphological and functional changes.
In contrast, in dynamic environments where cells are exposed to fluid flow, adhesion faces significant challenges. Shear forces influence the cell-surface connection, potentially leading to rearrangement of adhesion proteins and even cell detachment. In our experiments, we investigate how dynamic environments affect cell adhesion by applying various flow rates [2].
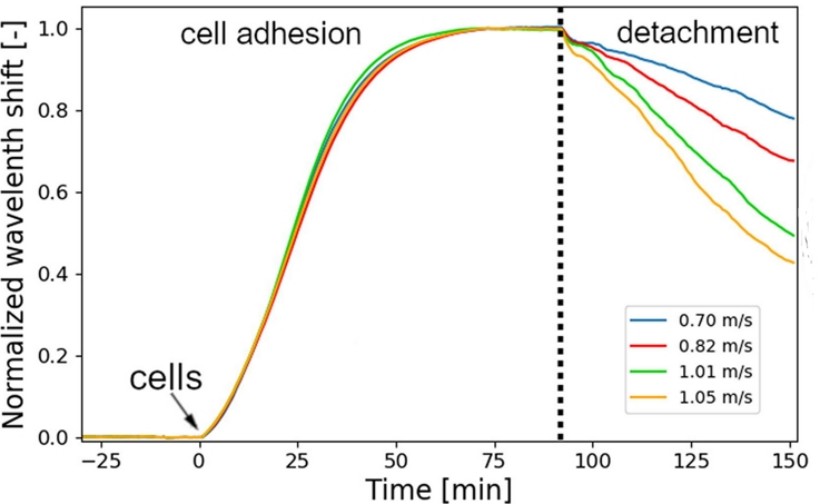
Figure 3: Representative cell adhesion curves and the characteristics of cell detachment induced by different flow rates [2].
Under lower shear forces, cells gradually adapt, while at higher shear forces, certain cells detach.
Figure 4: The cell detachment ratio from the 1800 RPM measurement in the function of the velocity and the fitted linear function [2].
This is particularly important for understanding vascular diseases and the metastasis of cancer cells, as mechanical forces play a significant role in these processes.
One of the goals of our research group is to understand the adhesion properties of cells in various environments, with particular attention to static and dynamic conditions and the effects of enzymatic modifications to the glycocalyx layer. Such research can contribute to a deeper understanding of cellular mechanobiology, and the development of new therapeutic strategies.
For studying cell adhesion, we use a set of novel techniques:
A. Optical biosensor measurements
Our group investigates cell adhesion with label-free optical biosensors in close collaboration with the HUN-REN EK MFA Nanobiosensorics Laboratory (www.nanobiosensorics.com), where several novel biophysical tools are available. These instruments allow for high throughput measurements without labelling molecules. Although this technique measures adhesion force only in an indirect manner, it is very sensitive to changes in the refractive index in a 200 nm thick surface layer above the sensor surfaces. This allows to study of the kinetics of different biological processes with an excellent time resolution and enables biophysical modelling [3]. Using label-free optical biosensors, we investigate the adhesion of individual cells and endothelial cell layers, as well as the effect of different molecules on the kinetics of cell adhesion.
Figure 5: Biosensor plate [4].

Figure 6: The working principle of optical biosensors. A laser light propagates in the optical waveguide inducing an evanescent field above the sensor surface where we can detect the adhered cells [5].
B. Fluid Force Microscopy measurements
We use fluid force microscopy (FluidFM), available at the Nanobiosensorics Laboratory at HUN-REN EK MFA (www.nanobiosensorics.com), to directly measure the adhesion force of a single cell at a certain time point. FluidFM uses a modified atomic force microscope cantilever with a microfluidic channel connected to a pressure controller system. If a negative pressure is generated inside the channel, a cell or microbead can attach to an aperture at the end of the cantilever, while overpressure can be used to quickly remove it [6].
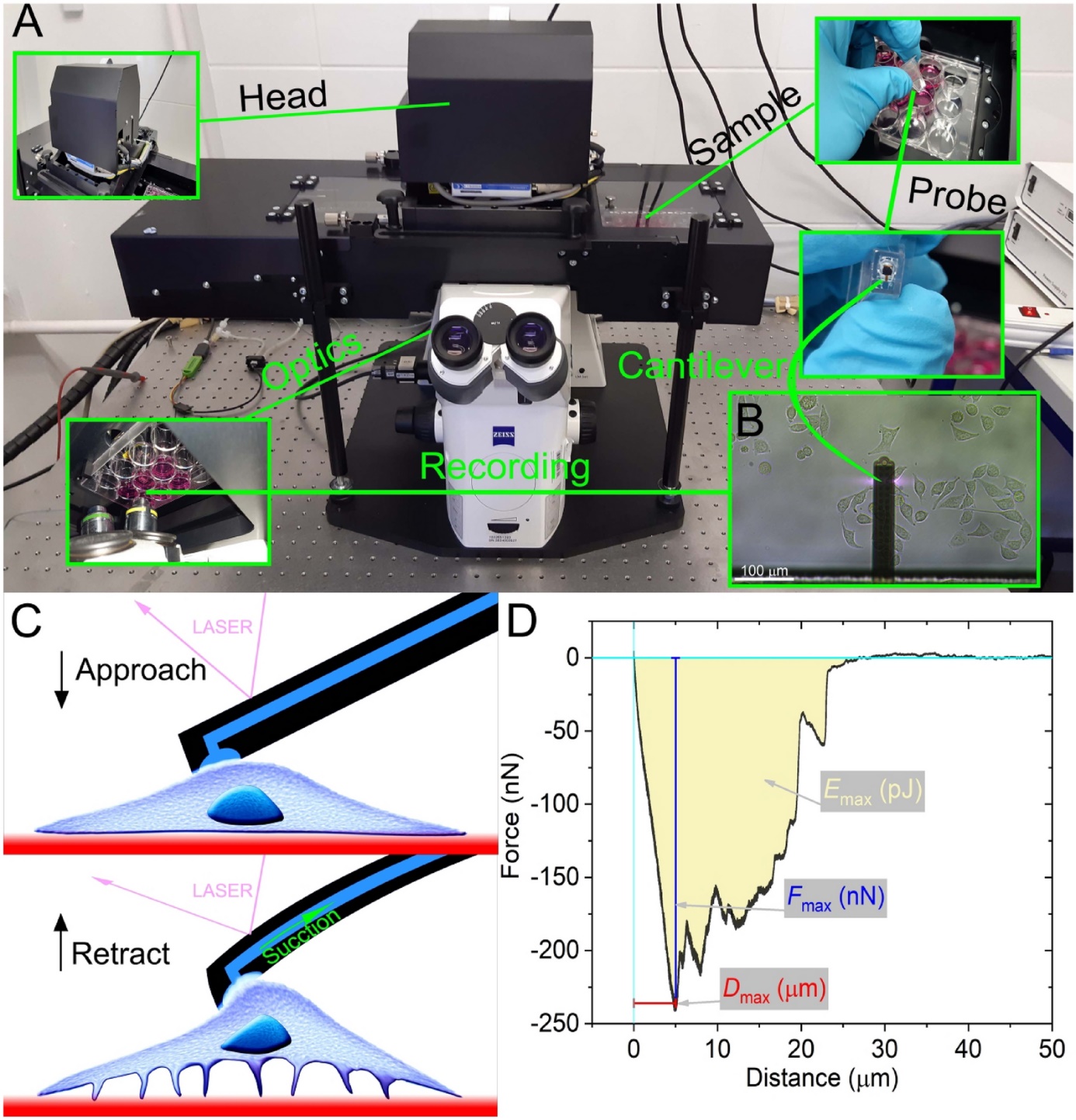
Figure 7: Procedure of adhesion measurements with FluidFM [6].
The same method is also used to investigate the strength of cell-cell interactions. In our work in collaboration with Prof. Mária Deli's Biological Barriers Research Group, we are treating endothelial cells with cARLA molecule cocktail [7], developing blood-brain barrier models that can be used to gain a more accurate picture of cerebrovascular disease and predict the entry of certain drug molecules into the brain.
Using precise pressure and time control, different molecules can be injected into the cytoplasm or nucleus of individual cells in a pico-Liter volume scale after penetration into the cell membrane [8]. Using FluidFM, a fluorescent dye can be injected into endothelial cells forming a single cell layer. Afterwards, the spreading of the injected molecules into the surrounding cells and its effect on cell adhesion can be investigated at different time points. In our collaboration work, we inject the cARLA cocktail into endothelial cells. Because previous diffusion-based experiments have shown that cARLA strengthens cell-cell contacts, our main aim is to study how the maturation of the blood-brain barrier alters communication between cells through gap junctions upon simultaneous injection of fluorescent dye and cARLA.
Figure 8: Injection into single cells using the FluidFM technology (www.cytosurge.com).
[1] Kanyo N, Kovacs KD, Saftics A, Szekacs I, Peter B, Santa-Maria AR, Walter FR, Dér A, Deli MA, Horvath R. Glycocalyx regulates the strength and kinetics of cancer cell adhesion revealed by biophysical models based on high resolution label-free optical data. Sci Rep. 2020 Dec 30;10(1):22422. doi: 10.1038/s41598-020-80033-6. PMID: 33380731; PMCID: PMC7773743.
[2] Kovács et al. Label-free tracking of whole-cell response on RGD functionalized surfaces to varied flow velocities generated by fluidic rotation, Journal of Colloid and Interface Science, Volume 599, 2021, Pages 620-630, ISSN 0021-9797, https://doi.org/10.1016/j.jcis.2021.04.091.
[3]Sztilkovics M, Gerecsei T, Peter B, Saftics A, Kurunczi S, Szekacs I, Szabo B, Horvath R. Single-cell adhesion force kinetics of cell populations from combined label-free optical biosensor and robotic fluidic force microscopy. Sci Rep. 2020 Jan 9;10(1):61. doi: 10.1038/s41598-019-56898-7. PMID: 31919421; PMCID: PMC6952389.
[4] https://nanobiosensorics.com
[5] Kanyo N, Kovács KD, Kovács SV, Béres B, Peter B, Székács I, Horvath R. Single-cell adhesivity distribution of glycocalyx digested cancer cells from high spatial resolution label-free biosensor measurements. Matrix Biol Plus. 2022 Feb 5;14:100103. doi: 10.1016/j.mbplus.2022.100103. PMID: 35243300; PMCID: PMC8857652.
[6] Ágoston G. Nagy, Nicolett Kanyó, Alexandra Vörös, Inna Székács, Attila Bonyár, Robert Horvath. Population distributions of single-cell adhesion parameters during the cell cycle from high-throughput robotic fluidic force microscopy. Sci Rep 12, 7747 (2022).
[7] G. Porkoláb, M. Mészáros, A. Szecskó, J.P. Vigh, F.R. Walter, R. Figueiredo, I. Kálomista, Z. Hoyk, G. Vizsnyiczai, I. Gróf, J. Jan, F. Gosselet, M.K. Pirity, M. Vastag, N. Hudson, M. Campbell, S. Veszelka, & M.A. Deli, Synergistic induction of blood–brain barrier properties, Proc. Natl. Acad. Sci. U.S.A. 121 (21) e2316006121, https://doi.org/10.1073/pnas.2316006121 (2024)
[8] Kovács, K. D., Visnovitz, T., Gerecsei, T., Peter, B., Kurunczi, S., Koncz, A., Németh, K., Lenzinger, D., Vukman, K. V., Balogh, A., Rajmon, I., Lőrincz, P., Székács, I., Buzás, E. I., & Horvath, R. (2023). Nanoinjection of extracellular vesicles to single live cells by robotic fluidic force microscopy. Journal of Extracellular Vesicles, 12, e12388. https://doi.org/10.1002/jev2.12388

senior research associate

senior research associate

PhD student

PhD student
 HORVÁTH Róbert
HORVÁTH Róbert
|
senior research associate | publications | CV |
 MARTINS Ana
MARTINS Ana
|
senior research associate | publications | CV |
 RAJMON Imola
RAJMON Imola
|
PhD student | publications | CV |
 VARGA Sebestyén
VARGA Sebestyén
|
PhD student | publications | CV |