
Recent technological advancements in systems biology, lab automation, and high-throughput microscopy have opened the door to systematic discovery of complex biological systems using high-throughput light microscopy. Modern equipments produce massive amounts of data which cannot be analyzed manually.
Automating the analysis process poses several challenges related to computational cell biology. Our group dedicated to finding computational solutions to biological problems. Our research focuses on the intersection of biology and computer science, and combine wet-lab and light microscopy with image analysis and machine learning methods.
Image analysis Machine learning Single cell analysis Imaging
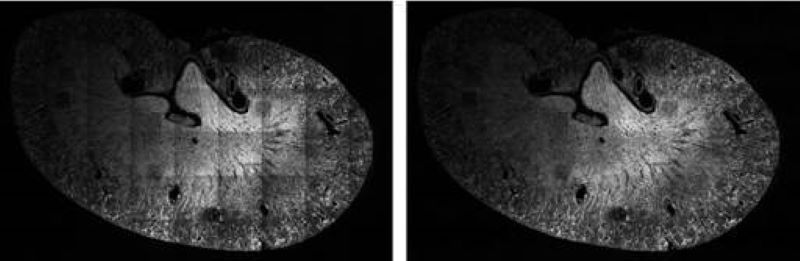
Illumination Correction
For quantitative measurements based on light microscopy and especially fluorescent intensities, it is essential to normalize the image data to correct for aberrations inherent in the acquisition process. One common source of error is the result of a non-ideal illumination field produced by the objective. Our novel algorithms addresses these issues using energy minimization. The corrected field resulting from our approach is extremely flat, and we can achieve this level of quality without requiring a calibrated reference sample.
Reconstruction
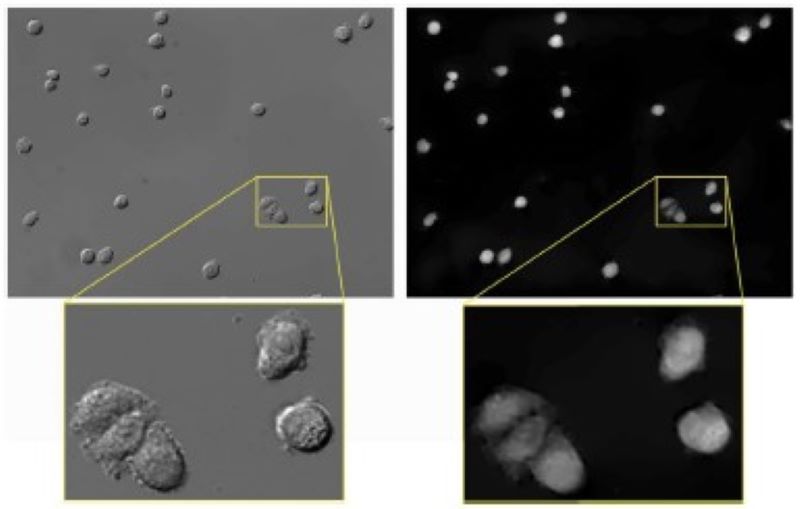
We developed an algorithm based on energy minimization to convert differential interference contrast (DIC) images to phase images to make them easier to analyze.
Tracking
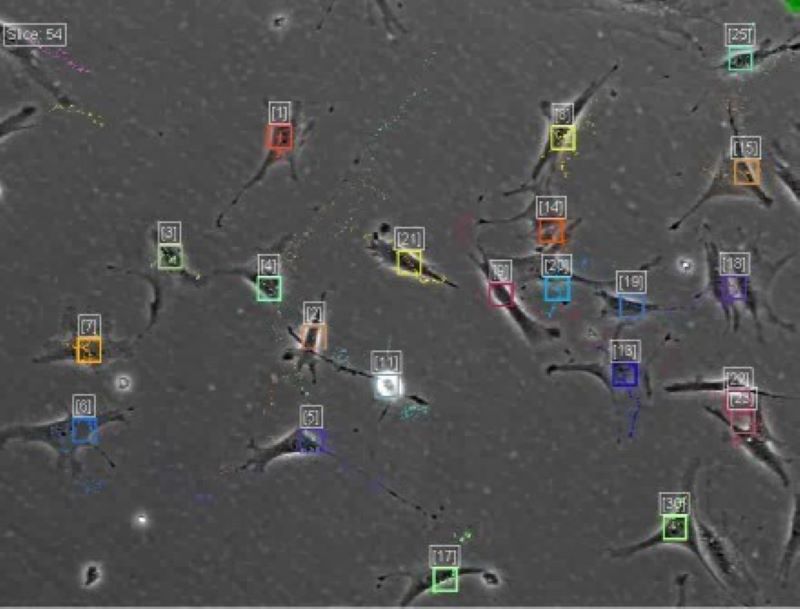
We are interested in developing methods for identifying and tracking cells or sub-cellular structures on live cell images. We have been developed a software the CellTracker, which corrects illumination problems, finds alignments, as well as automatically and manually tracks cells, mainly on phase contrast images. The program is available with MATLAB GUI. [Download CellTracker]
Segmentation of overlapping cells (the 'gas of circles' model)
Variational methods for shape modeling to extract near-circular objects (e.g. nuclei).
The multi-layered 'gas of near-circles' model is capable of segmenting touching or even overlapping cells on high confluency images.
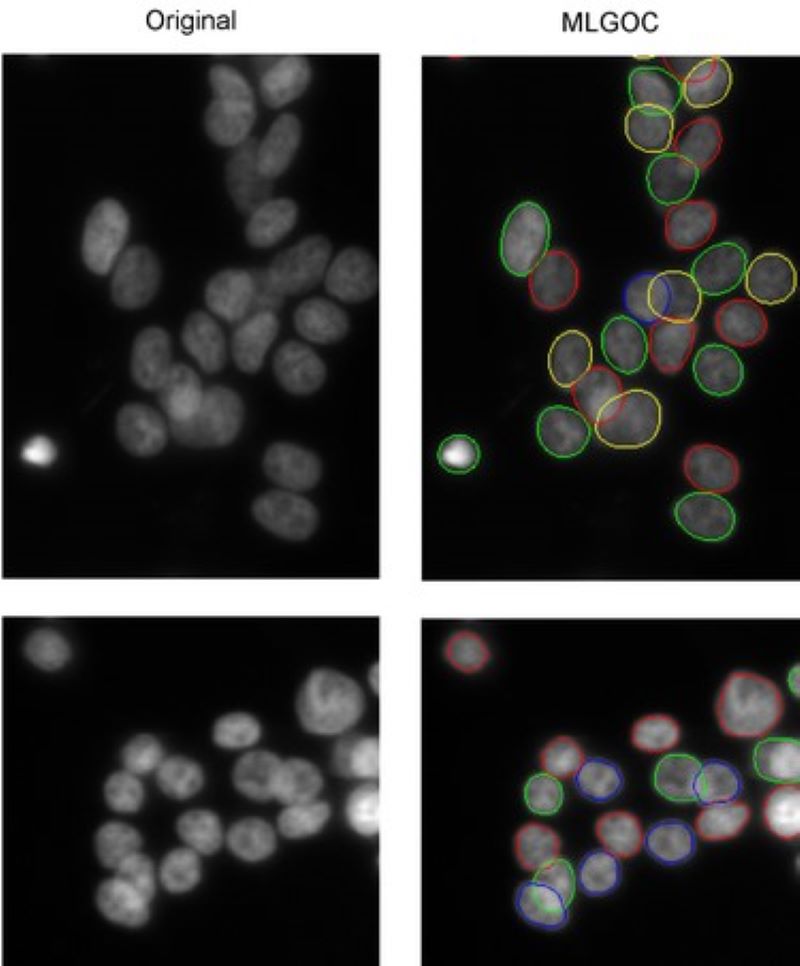
Selective Active Contours
The selective active contours utilize simple shape characteristics such as area and perimeter, to describe objects that can provide computationally efficient shape selective segmentation.
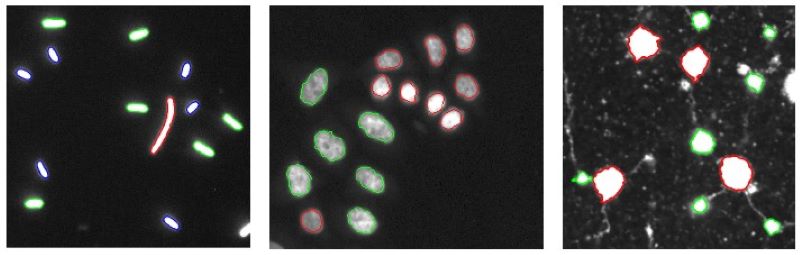
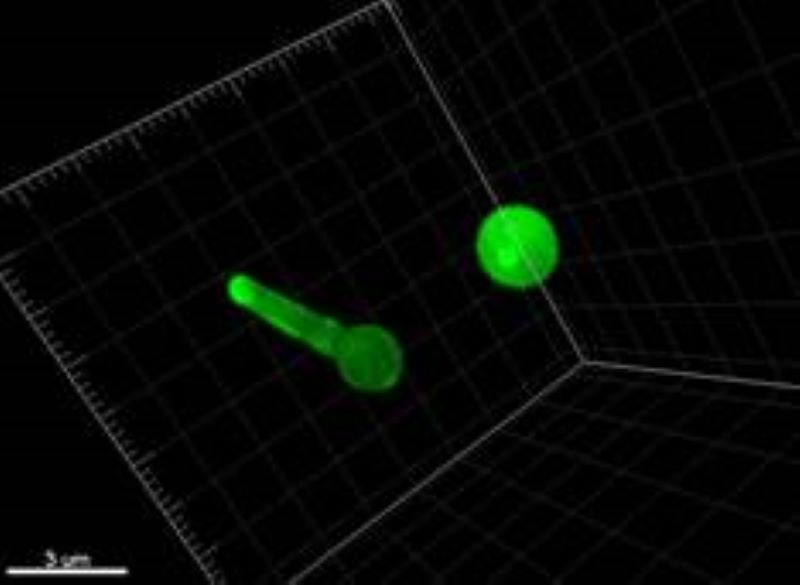
Selective Active Contours in 3D
3D extension of the selective 2D active contours.

Cells in pseudohyphae and in normal form. Finding the pseudohyphae form.
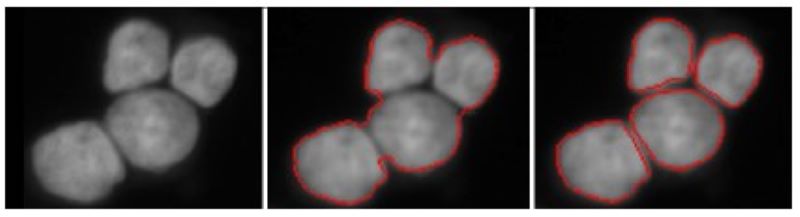
Spliting touching cells
Segment individual cell nuclei by splitting touching ones. The two-step approach merely based on energy minimization principles using a higher-order active contour framework.
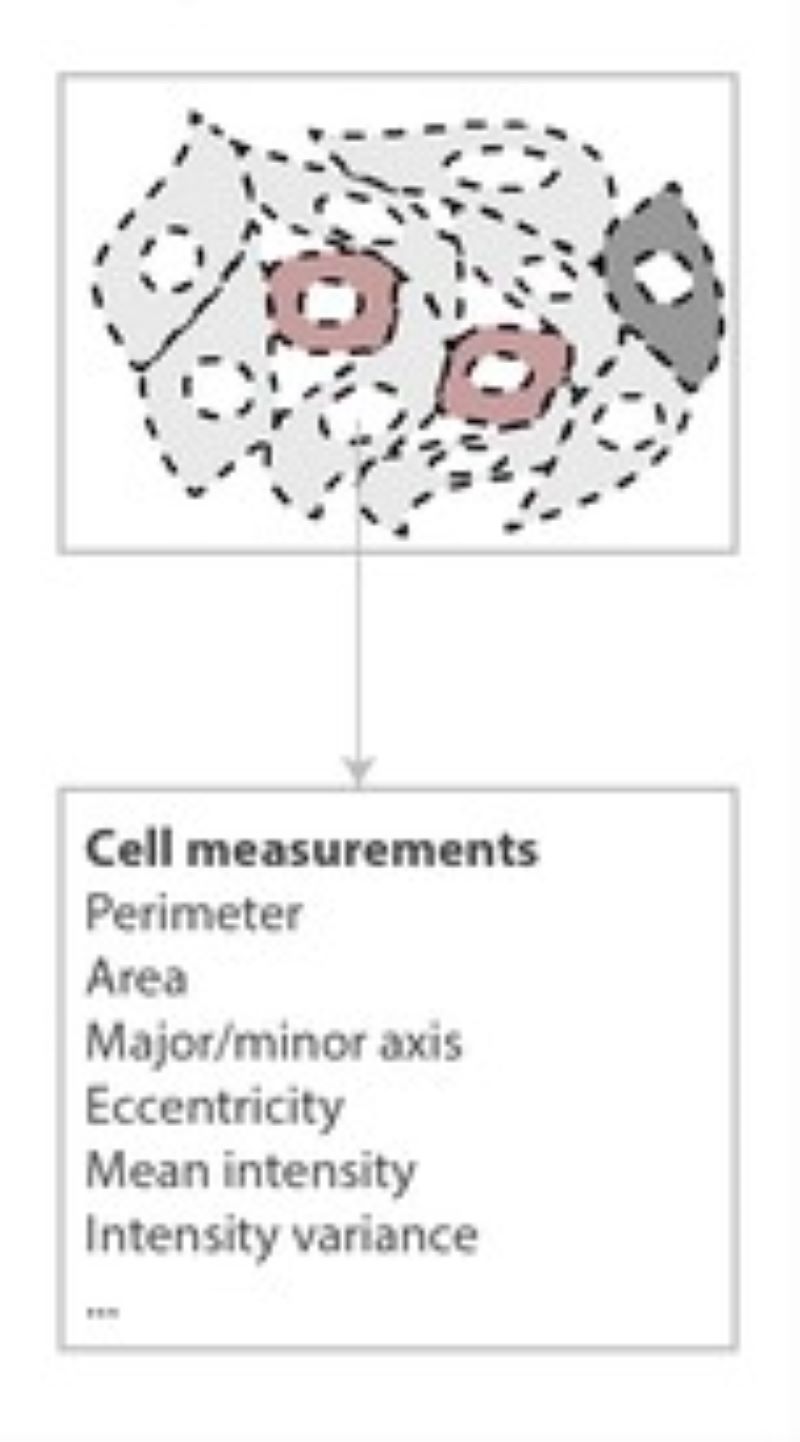
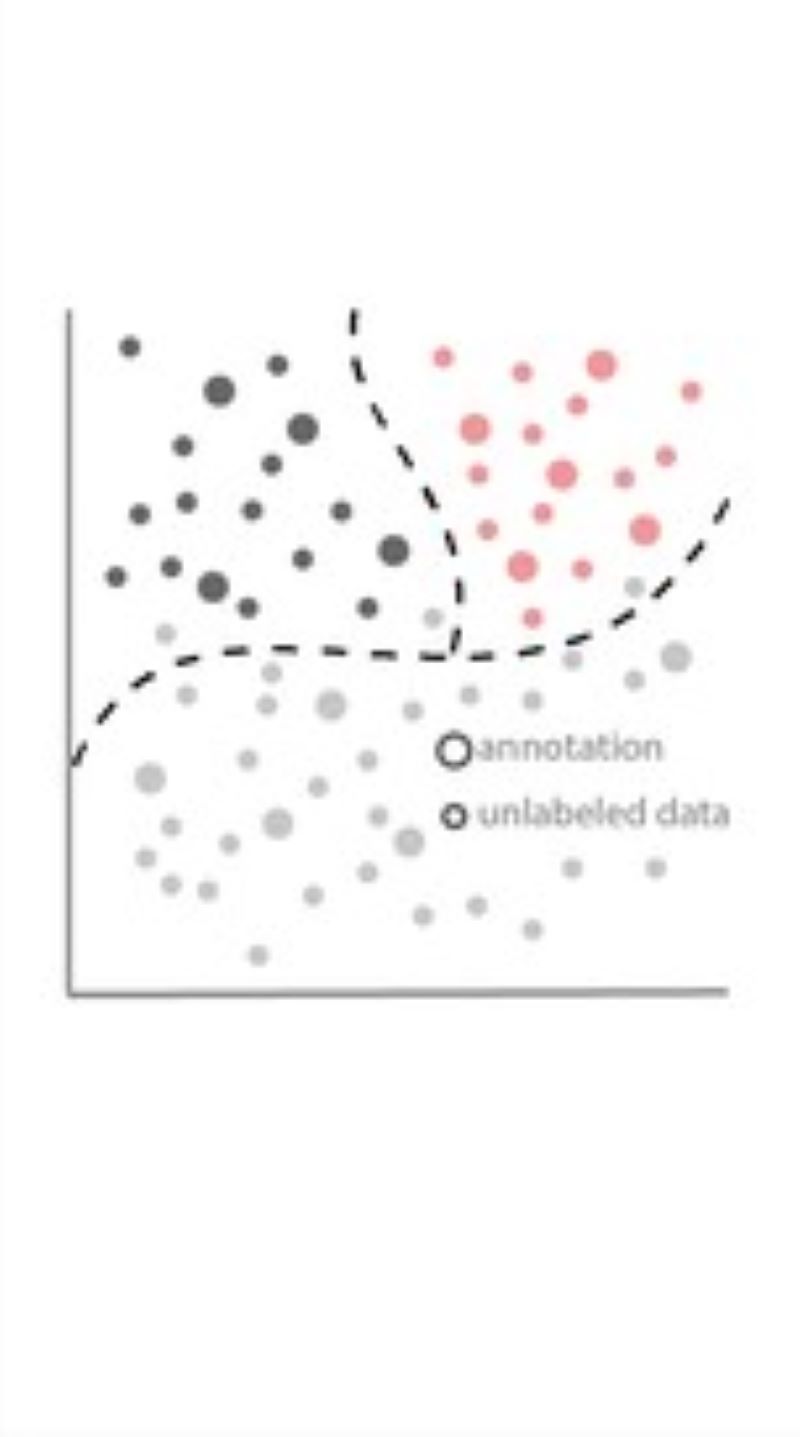
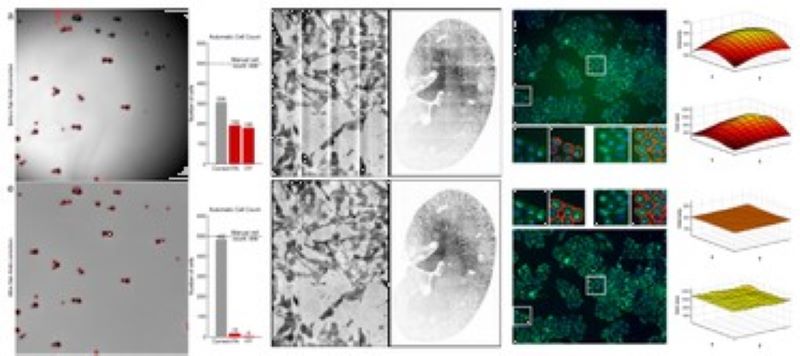
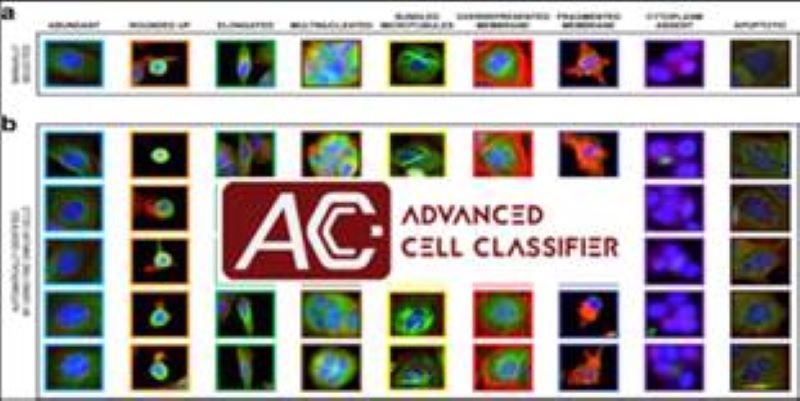
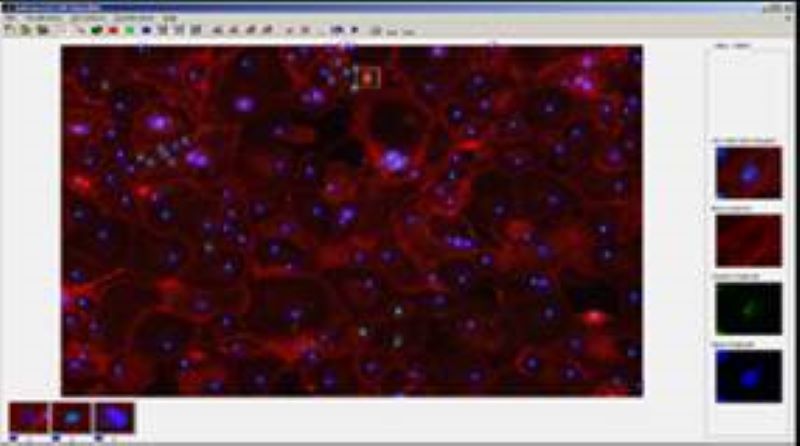
Phenotyping - Advanced Cell Classifier
Advanced Cell Classifier is a data analyzer program to evaluate cell-based high-content screens and tissue section images developed at the Biological Research Centre, Szeged and FIMM, Helsinki (formerly at ETH Zurich). The basic aim is to provide a very accurate analysis with minimal user interaction using advanced machine learning methods. ACC was used to analyze some of the first large whole genome scale RNAi screens and all together for more than 300.000.000 images and several billion single cell-based machine learning decisions.
Deep Learning
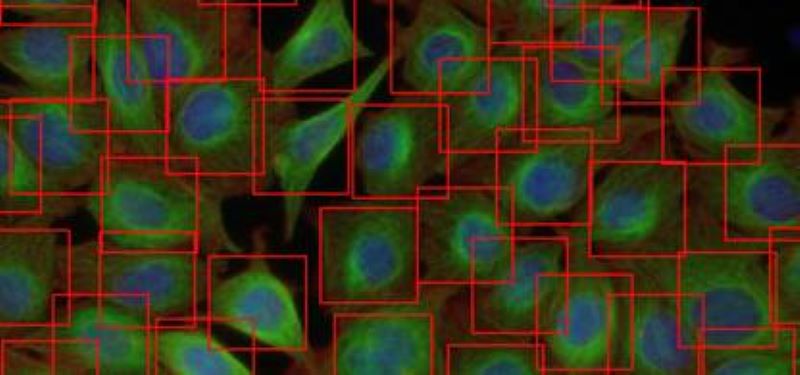
We developed a fast and fully automated tools that assesses the number and location of cells using Deep Convolutional Neural Networks (DCNN). Our methods highly outperforms state-of-the-art machine learning models and provides comparable detection accuracy to human field experts.
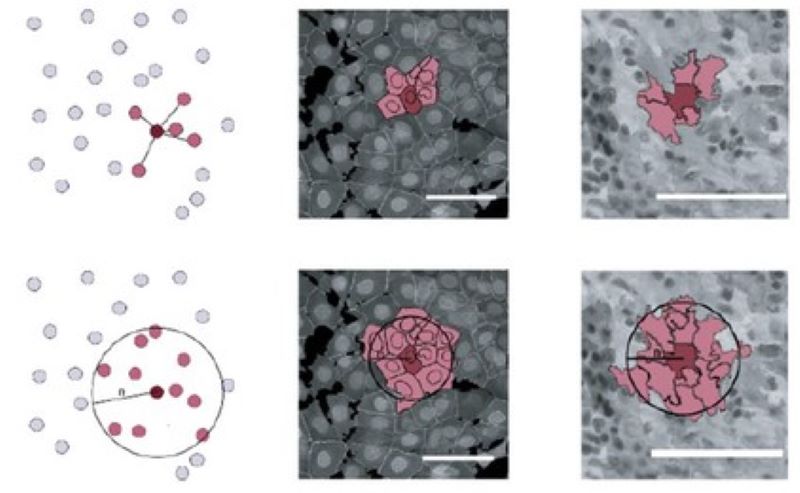
Microenvironment-based phenotyping
We research how various microenvironmental features contribute to identifying a cell and how such additional information can improve single-cell-level phenotypic image analysis.
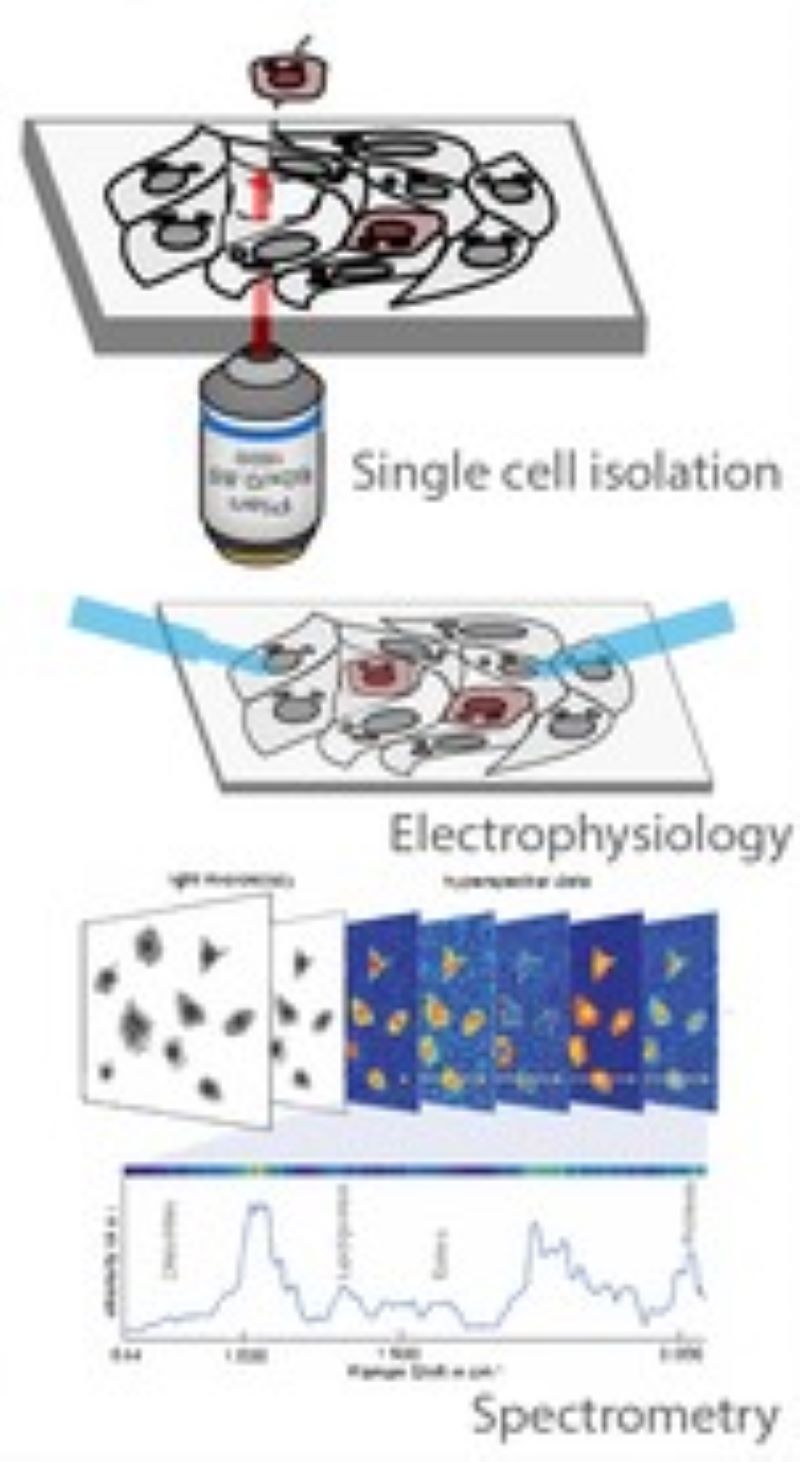
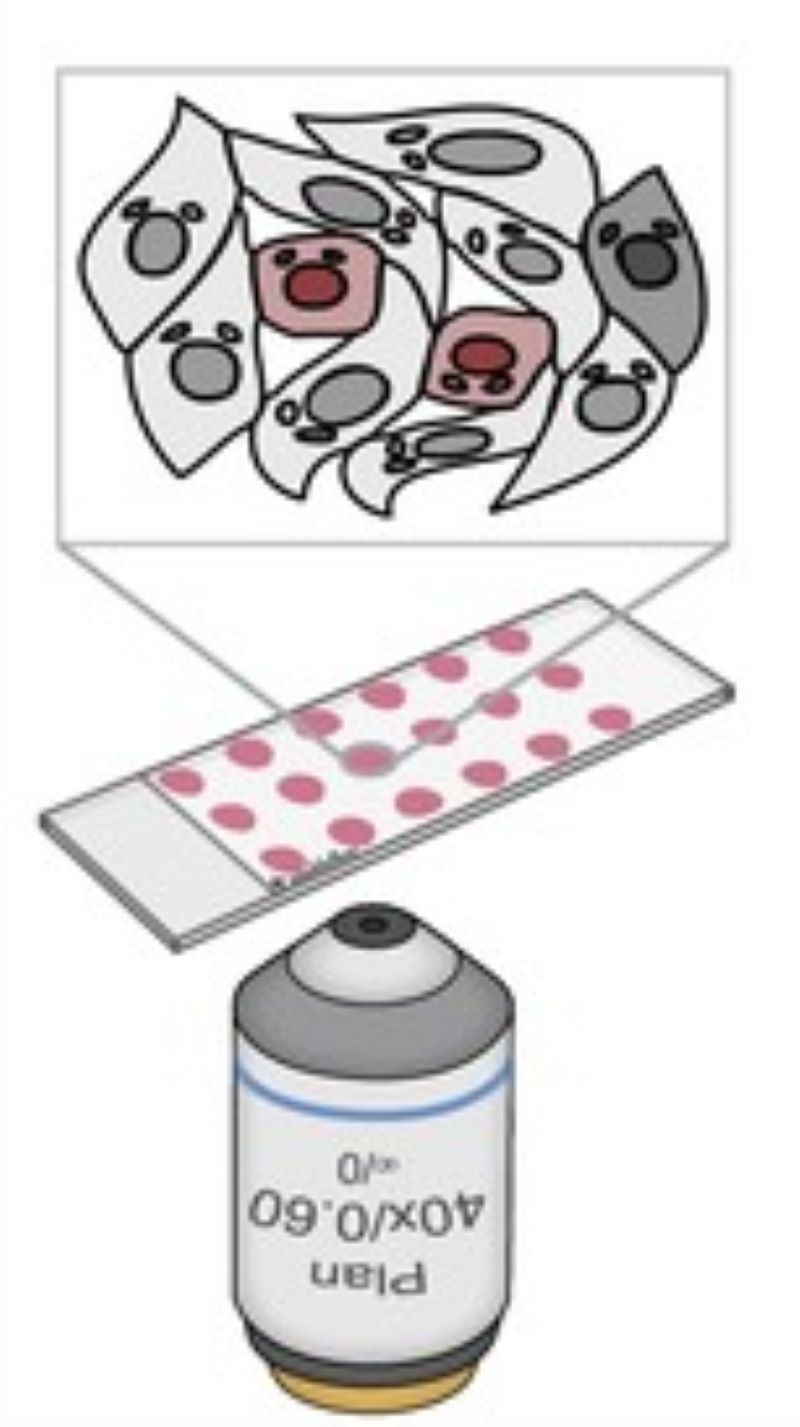
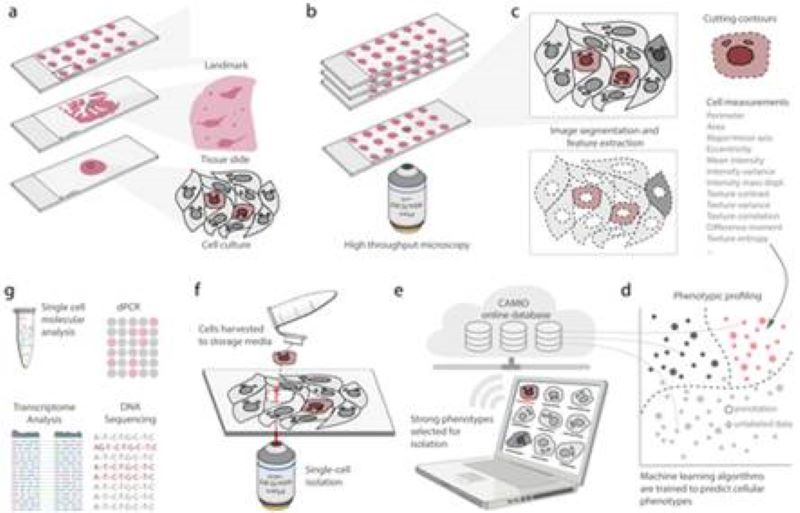
CAMI - Computer Aided Microscopy Isolation system
We develop a high-throughput, non-disruptive, and cost-effective isolation methods that is capable of capturing individually targeted cells using widely available techniques. Using high-resolution microscopy, laser microcapture microscopy, image analysis, and machine learning, our technology enables scalable molecular genetic analysis of single cells, targetable by morphology or location within the sample. Cell data along with the location and contour ofeach cell is sent to our interactive online database CAMIO.
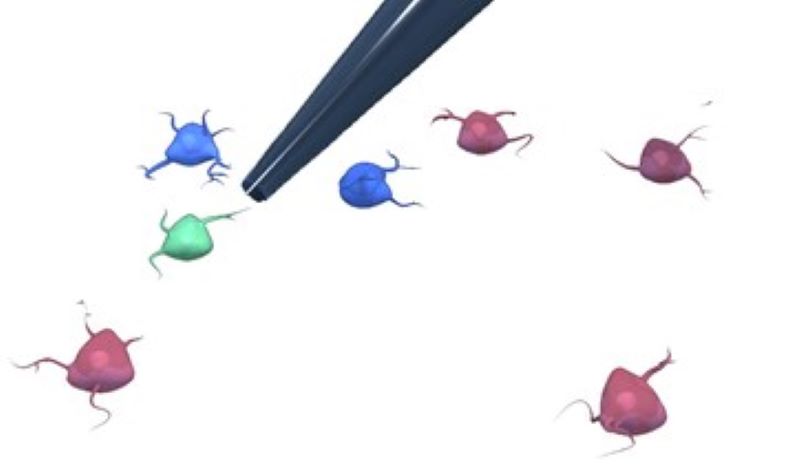
AutoPatcher
We are building an automated patch clamp system to analyze the electrophysiological properties of neurons in vitro. The system automatically selects a cell in label-free microscopy and performs patch clamping on it using image processing and deep learning.
High content screening
The HCS technology employs different automated microscopes in a high throughput format to extract quantitative information from cells or tissue samples based on various parameters such as spatial distribution or the morphology changes of the target cells. To address these various cellular phenotypes, both widefield and confocal microscopes are used in the BIOMAG group:
PerkinElmer Operetta
Main features:
Analysis of cellular phenotypes:

Leica SP8-digital light sheet

Laser microdissection systems
During LMD, a laser is focused on the tissue and it cuts the sample alongside a predefined trajectory. After the cutting process, the required elements can be extracted and collected for further analysis. The dissected material is then available for further downstream applications such as genomics, transcriptomics, next generation sequencing, proteomics or other analytical techniques. Based on the movement of the laser and sample collection, two main approaches have been used:
Leica LMD6
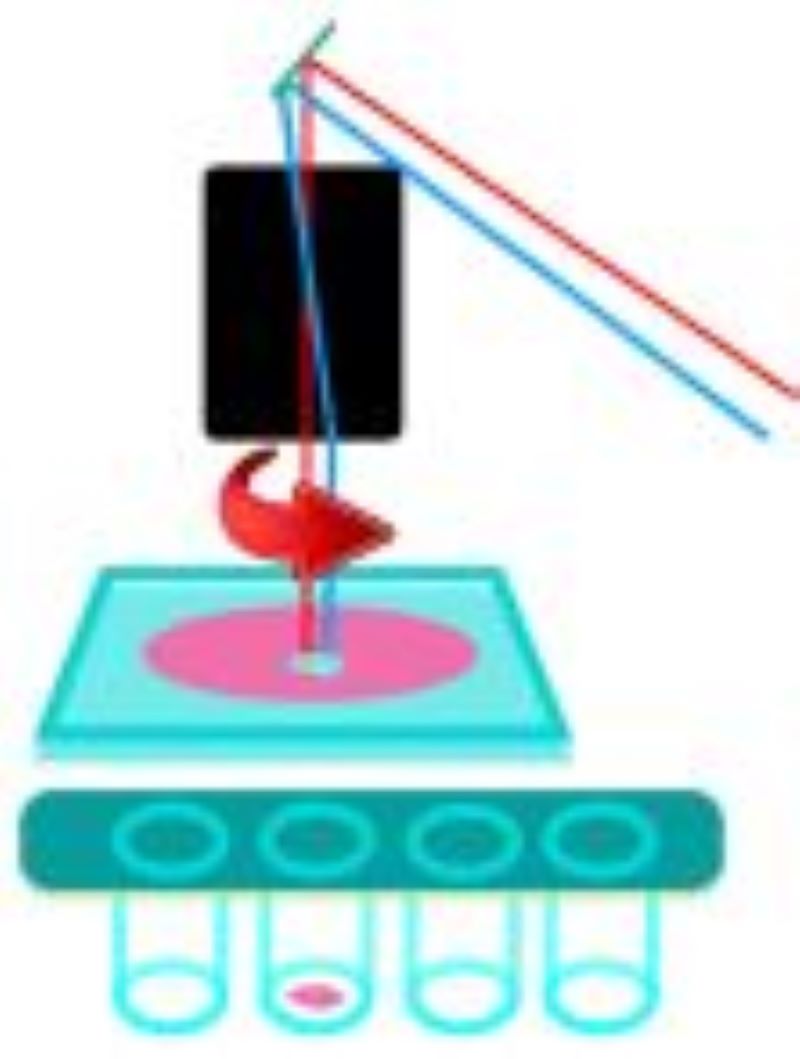
Zeiss Palm Microbeam
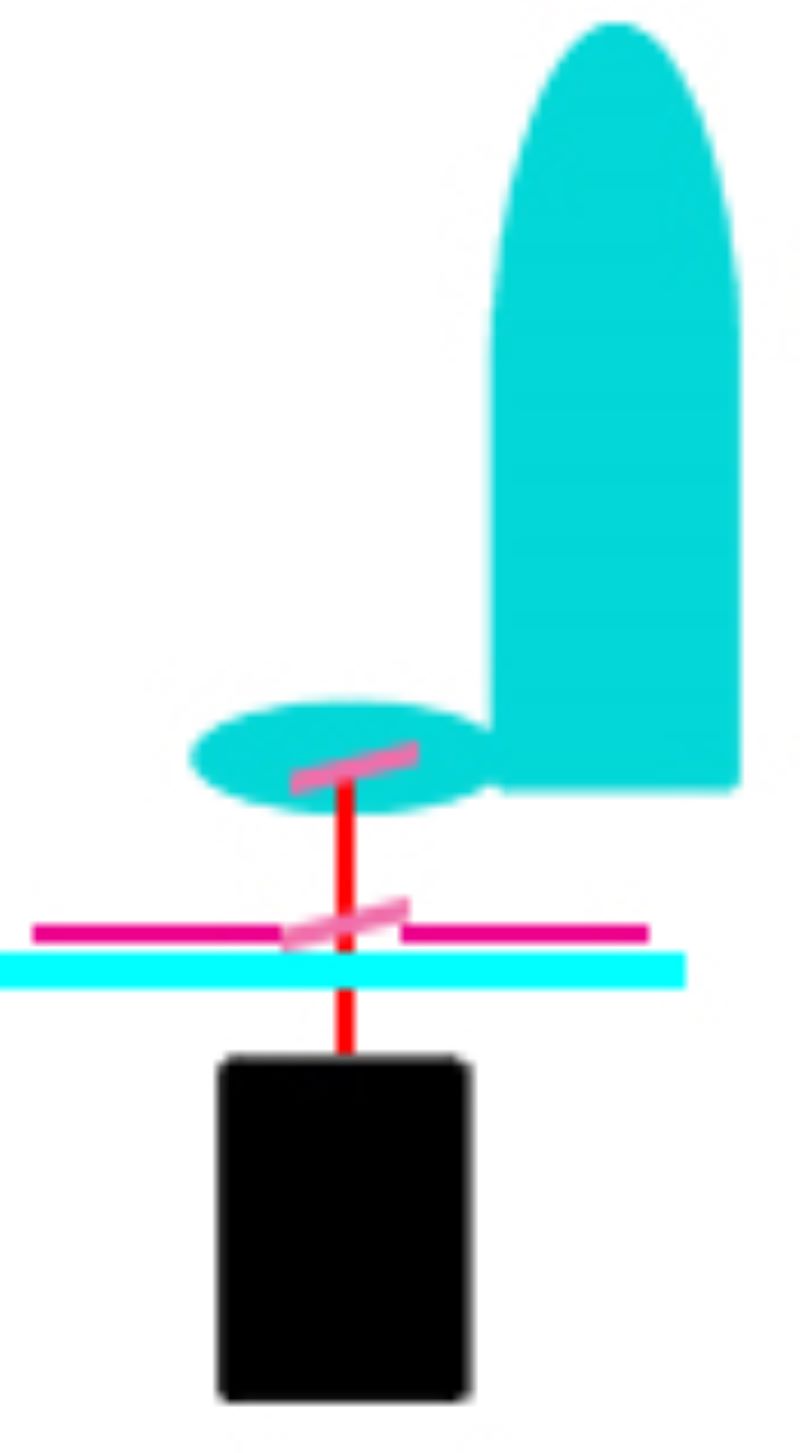

Senior Research Associate

Research Associate

Research Associate

Research Associate

Research Associate

Research Associate

Research Associate

Junior Research Associate

Junior Research Associate

Scientific Administrator

Junior Research Associate

Scientific Administrator
Junior Research Associate

PhD Student

Scientific Administrator

Scientific Administrator

Scientific Administrator

Scientific Administrator

Laboratory Assistant
 Péter HORVÁTH
Péter HORVÁTH
|
Senior Research Associate | publications | CV |
 Krisztina BUZÁS
Krisztina BUZÁS
|
Research Associate | publications | CV |
 Ede MIGH
Ede MIGH
|
Research Associate | publications | |
 Vivien CSAPÓNÉ MICZÁN
Vivien CSAPÓNÉ MICZÁN
|
Research Associate | publications | |
 Edina GYUKITY-SEBESTYÉN
Edina GYUKITY-SEBESTYÉN
|
Research Associate | publications | |
 Mária HARMATI
Mária HARMATI
|
Research Associate | publications | CV |
 Gabriella GRESKOVICS-DOBRA
Gabriella GRESKOVICS-DOBRA
|
Research Associate | publications | |
 Ákos DIÓSDI
Ákos DIÓSDI
|
Junior Research Associate | publications | |
 István GREXA
István GREXA
|
Junior Research Associate | publications | |
 Ervin TASNÁDI
Ervin TASNÁDI
|
Scientific Administrator | publications | |
 Viktor PÁL
Viktor PÁL
|
Junior Research Associate | ||
 Zsanett Zsófia IVÁN
Zsanett Zsófia IVÁN
|
Scientific Administrator | ||
 Mátyás BUKVA
Mátyás BUKVA
|
Junior Research Associate | publications | CV |
 Tímea BÖRÖCZKY
Tímea BÖRÖCZKY
|
PhD Student | publications | CV |
 Dávid BAUER
Dávid BAUER
|
Scientific Administrator | ||
 Ferenc KOVÁCS
Ferenc KOVÁCS
|
Scientific Administrator | ||
 András KRISTON
András KRISTON
|
Scientific Administrator | publications | |
 Lilla PINTÉR
Lilla PINTÉR
|
Scientific Administrator | ||
 Nóra HAPEK
Nóra HAPEK
|
Laboratory Assistant |