
We study how molecular networks and genomes evolve by combining computational and high-throughput experimental approaches.
Our research is dominantly focused on three main topics:
1) Evolution of metabolism
Metabolism is central to life as it provides the building blocks and energy for all biological processes. While their fundamental tasks are highly conserved across all life forms, metabolic networks differ across species both in their pathway composition and in the quantitative details of how they work. We study both aspects of metabolic evolution by focusing on two basic questions: (i) How do new metabolic pathways evolve? The prevailing view is that evolution capitalizes on the weak side activities of preexisting enzymes. We systematically investigate how such ‘underground reactions’ contribute to the utilization of new nutrients and to the synthesis of industrially important compounds. (ii) What are the general principles governing the evolution of metabolite concentrations? By comparing metabolomes of different species, we ultimately aim to understand which metabolite differences matter for health.
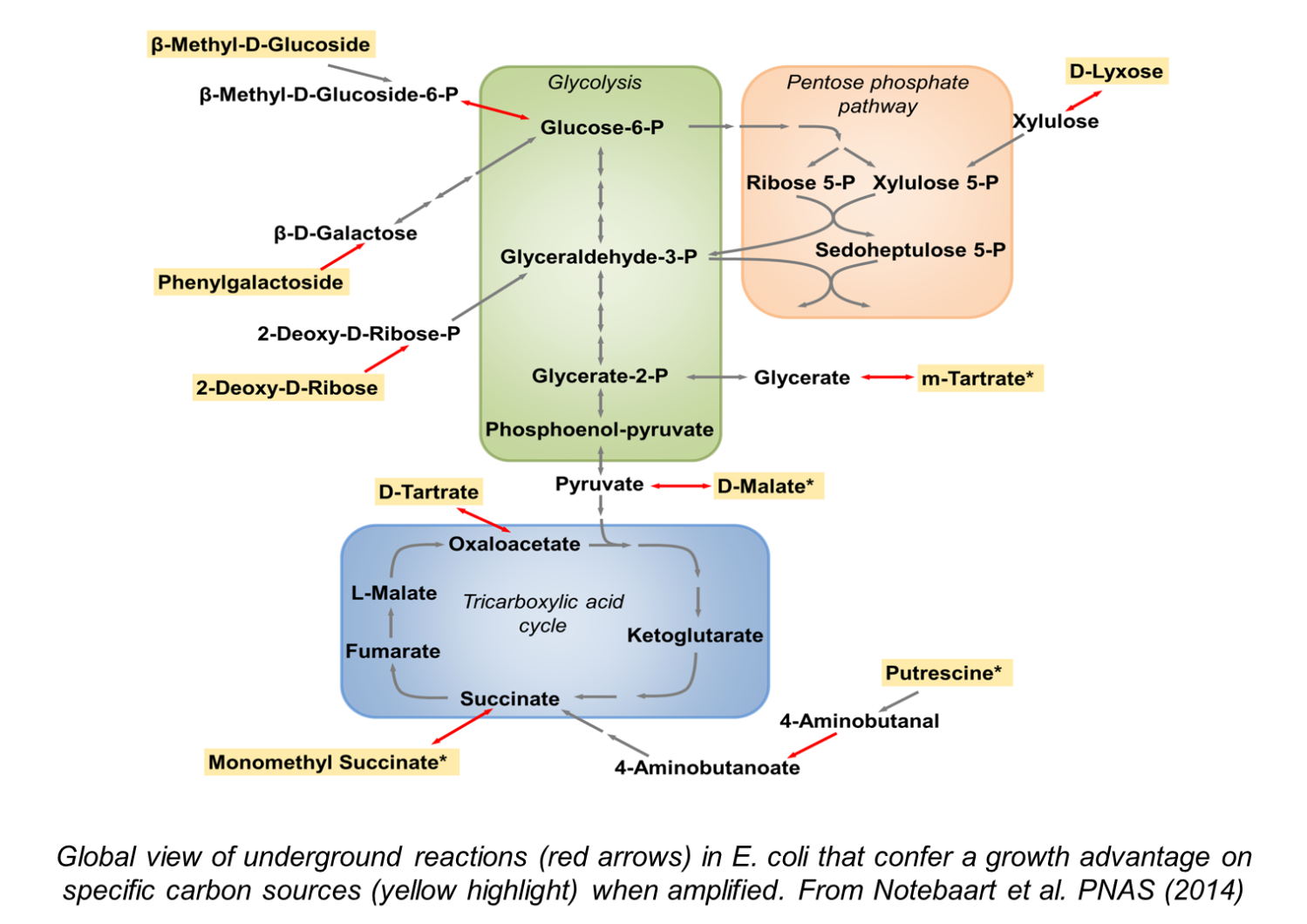
2) Mobility of antimicrobial resistance genes
Horizontal gene transfer between bacterial lineages is widespread and play a key role in the evolution of antimicrobial resistance. Despite its clinical importance, however, we have only a limited understanding of (i) the general trends and impacts of gene exchange between virulent pathogens and multi-drug resistant commensal bacteria, and (ii) how genes conferring resistance to different classes of antimicrobial drugs vary in their mobilization potenital. We address these issues by analyzing the gene exchange networks of human microbiota, multi-drug resistant and pathogenic bacteria alike.
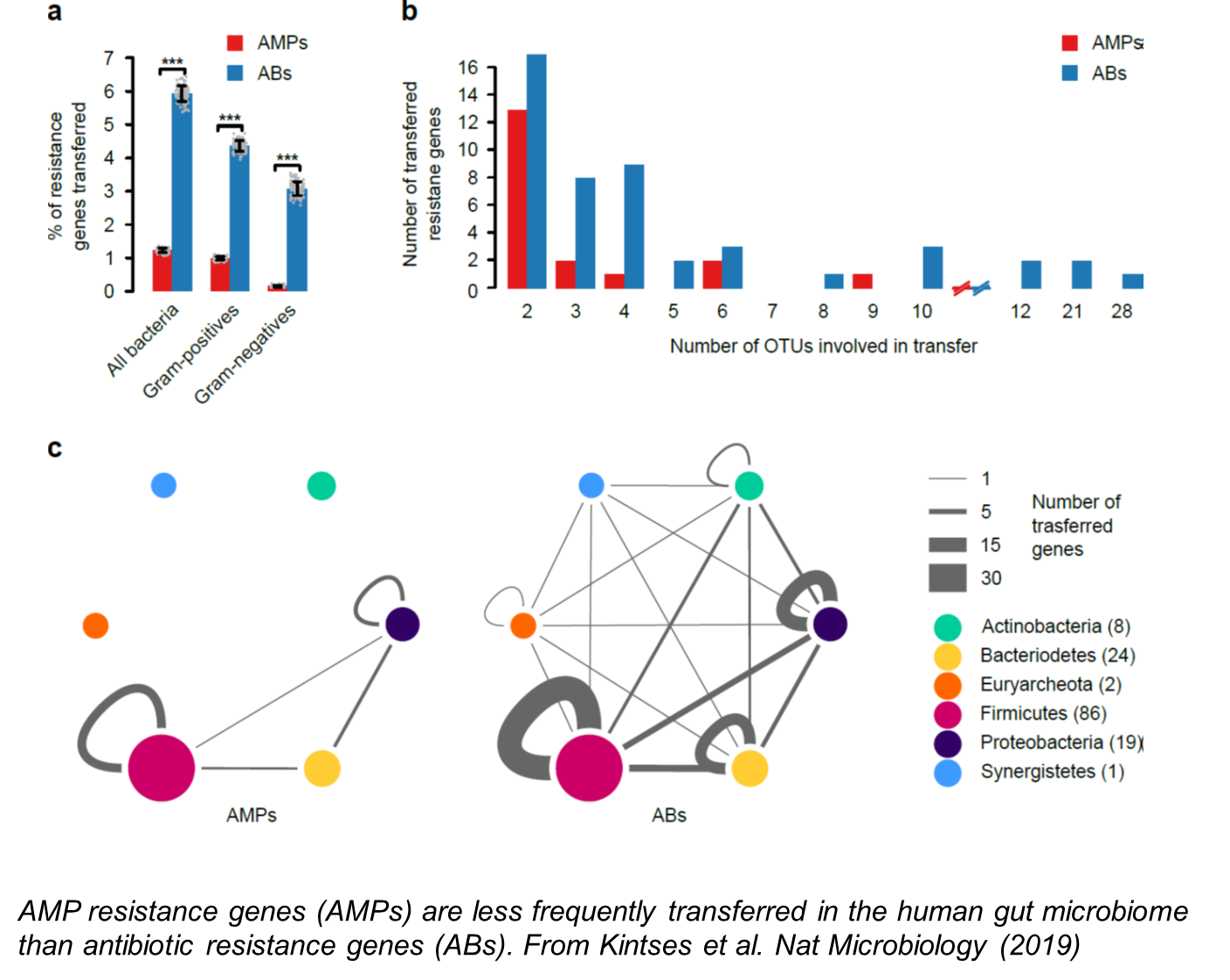
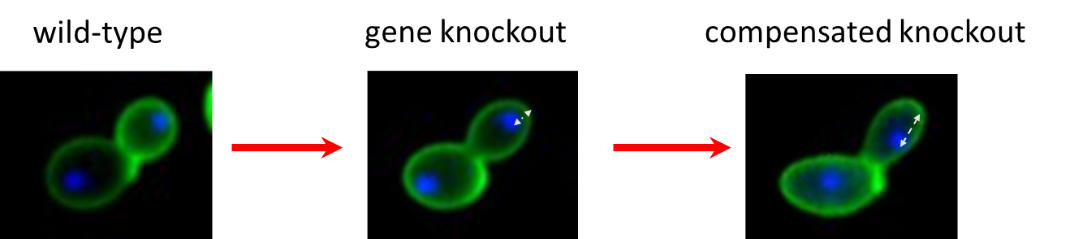
3) Compensatory evolution and the constructive role of harmful mutations
What are the evolutionary forces driving phenotypic diversity? Traditional explanations include beneficial mutations, leading to adaptive changes, and neutral mutations. Our lab examines whether harmful mutations can also contribute to phenotypic diversity. As harmful mutations can be compensated by specific mutations elsewhere in the genome, such compensatory mutations may lead to divergence in phenotypic traits without direct selection on them. We systematically test this idea using laboratory evolution in budding yeast and E. coli and by focusing on the evolution of cellular morphology and metabolic traits, respectively.

senior research fellow

research fellow

research fellow

research fellow

research fellow

scientific administrator

scientific and technical administrator

scientific and technical administrator

scientific and technical administrator

scientific administrator

PhD student

laboratory assistant

MSc student

BSc student
 Balázs PAPP
Balázs PAPP
|
senior research fellow | publications | CV |
 Károly KOVÁCS
Károly KOVÁCS
|
research fellow | publications | |
 Balázs SZAPPANOS
Balázs SZAPPANOS
|
research fellow | publications | |
 Eszter ARI
Eszter ARI
|
research fellow | publications | CV |
 Zsuzsa SARKADI
Zsuzsa SARKADI
|
research fellow | publications | CV |
 Gergely FEKETE
Gergely FEKETE
|
scientific administrator | publications | |
 Gábor GRÉZAL
Gábor GRÉZAL
|
scientific and technical administrator | publications | |
 Orsolya LISKA
Orsolya LISKA
|
scientific and technical administrator | publications | |
 Ágoston HUNYA
Ágoston HUNYA
|
scientific and technical administrator | ||
 Anett NAGY-DEMCSÁK
Anett NAGY-DEMCSÁK
|
scientific administrator | publications | |
 Tamás STIRLING
Tamás STIRLING
|
PhD student | publications | |
 Fanni BIRTYIK
Fanni BIRTYIK
|
laboratory assistant | publications | |
 Lilla SZÉLES
Lilla SZÉLES
|
MSc student | ||
 István MAGYARY
István MAGYARY
|
BSc student |