
Our laboratory is committed to advancing precision medicine strategies to combat antibiotic resistance, a critical global health issue. Using cutting-edge systems and synthetic biology approaches—including bioinformatics, genome engineering, genomics and metagenomics—we aim to:
Ultimately, our work aims to serve as a guide on how modern biological approaches can be harnessed to develop personalized treatments capable of tackling even the most resilient bacteria.
The establishment of our laboratory was supported by the National Biotechnology Laboratory and a grant from the Hungarian Centre of Excellence for Molecular Medicine (HCEMM). HCEMM is dedicated to molecular medicine-oriented research initiatives, with the goal of fostering positive changes in research culture and promoting regional development.
Recent Media Coverage
Our work recently published in Cell on guiding phage therapy with genomic surveillance has recently attracted media attention. We’re excited to see this coverage bring attention to the urgent need for innovative treatments against antibiotic resistance.
For more details, please see here:
Job openings
We are seeking Postdocs and PhD students to join us. Contact Bálint Kintses if interested.
Global Mapping and Precision Phage Therapy for Multidrug-Resistant Acinetobacter baumannii Cell, 2024
In our recent publication in Cell, we created a comprehensive global map detailing the genetic diversity and geographical distribution of multidrug-resistant Acinetobacter baumannii, one of the most challenging nosocomial pathogens. By combining large-scale phylogeographical analysis with high-throughput phage typing, we analyzed an international genomic dataset from hospital samples across Eastern Europe and global public databases. Our study revealed that a few strain types dominate infections in each world region, and their geographical distribution remains stable over at least six years. This map facilitates the development of region-specific bacteriophage therapies, addressing a significant hurdle in phage therapy—the narrow host range of bacteriophages. Traditional phage therapeutics are limited by their specificity to certain bacterial strains, necessitating precision approaches for effective treatment. By identifying the dominant strains in specific regions, we can preemptively prepare phage collections that target the majority of local infections.
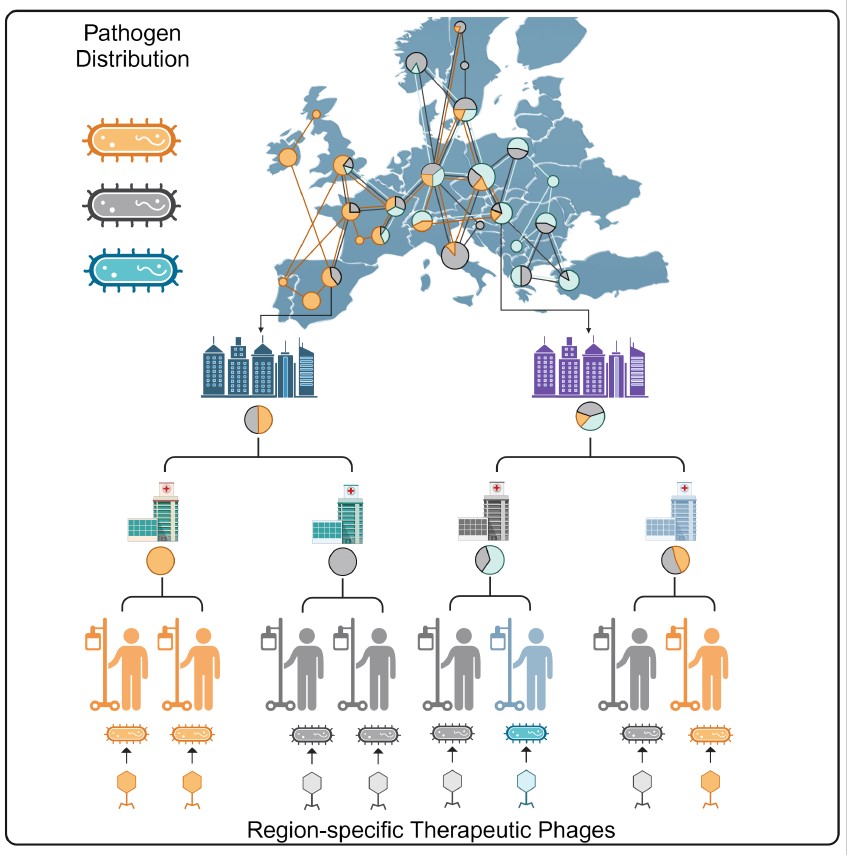
Key Findings:
To help visualize the mapping of bacterial variants and personalized phage therapy, this short animated video vividly illustrates the principles and significance of the research.
Reprogrammed Bacteriophage Particle Assisted Multi-species Functional Metagenomics (DEEPMINE)
In our publication in Nature Microbiology, we introduced a new approach called Reprogrammed Bacteriophage Particle Assisted Multi-species Functional Metagenomics (DEEPMINE) to expand the functional metagenomic toolbox for studying microbial communities and by doing so, deepen our understanding of how antimicrobial resistance emerges. The traditional metagenomic screening approach for antibiotic resistance genes (ARGs) relies on laboratory strains of Escherichia coli as bacterial hosts, leaving us blind to ARGs that provide resistance specifically in clinically relevant pathogens. To address this issue, DEEPMINE employs modified bacteriophage transducing particles to deliver large metagenomic plasmid libraries into a wide range of bacterial species, allowing us to perform metagenomic screens in clinically relevant bacterial pathogens from the Enterobacteriaceae family. Through this approach, we identified several previously unreported ARGs with species-specific effects on antibiotic susceptibility. Additionally, our study revealed that newly approved or late-stage clinical development antibiotics are just as prone to resistance formation as old antibiotics after decades of clinical use, highlighting the need for innovation and ongoing research into combating antibiotic resistance.
Analyzing the SARS-CoV-2 Epidemic Waves in Hungary
In our paper, we showed that retrospective evaluation of past waves of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic is key for designing optimal interventions against future waves and novel pandemics. Here, we reported on analyzing genome sequences of SARS-CoV-2 from the first two waves of the epidemic in 2020 in Hungary, mirroring a suppression and a mitigation strategy, respectively. Our analysis revealed that the two waves markedly differed in viral diversity and transmission patterns. Specifically, unlike in several European areas or in the USA, we found no evidence for early introduction and cryptic transmission of the virus in the first wave of the pandemic in Hungary. Despite the introduction of multiple viral lineages, extensive community spread was prevented by a timely national lockdown in March 2020. In sharp contrast, the majority of the cases in the much larger second wave can be linked to a single transmission lineage of the pan-European B.1.160 variant. This lineage was introduced unexpectedly early, followed by a 2-month-long cryptic transmission before a soar of detected cases in September 2020. Epidemic analysis revealed that the dominance of this lineage in the second wave was not associated with an intrinsic transmission advantage. This finding was further supported by the rapid replacement of B.1.160 by the alpha variant (B.1.1.7) that launched the third wave of the epidemic in February 2021. These findings collectively underscore the role of the founder effect, in conjunction with cryptic transmission, as the driving factors behind viral diversity, as opposed to recurrent international introductions or heightened transmissibility.
Evaluating the Risk of Resistance Evolution Against Novel Antimicrobials
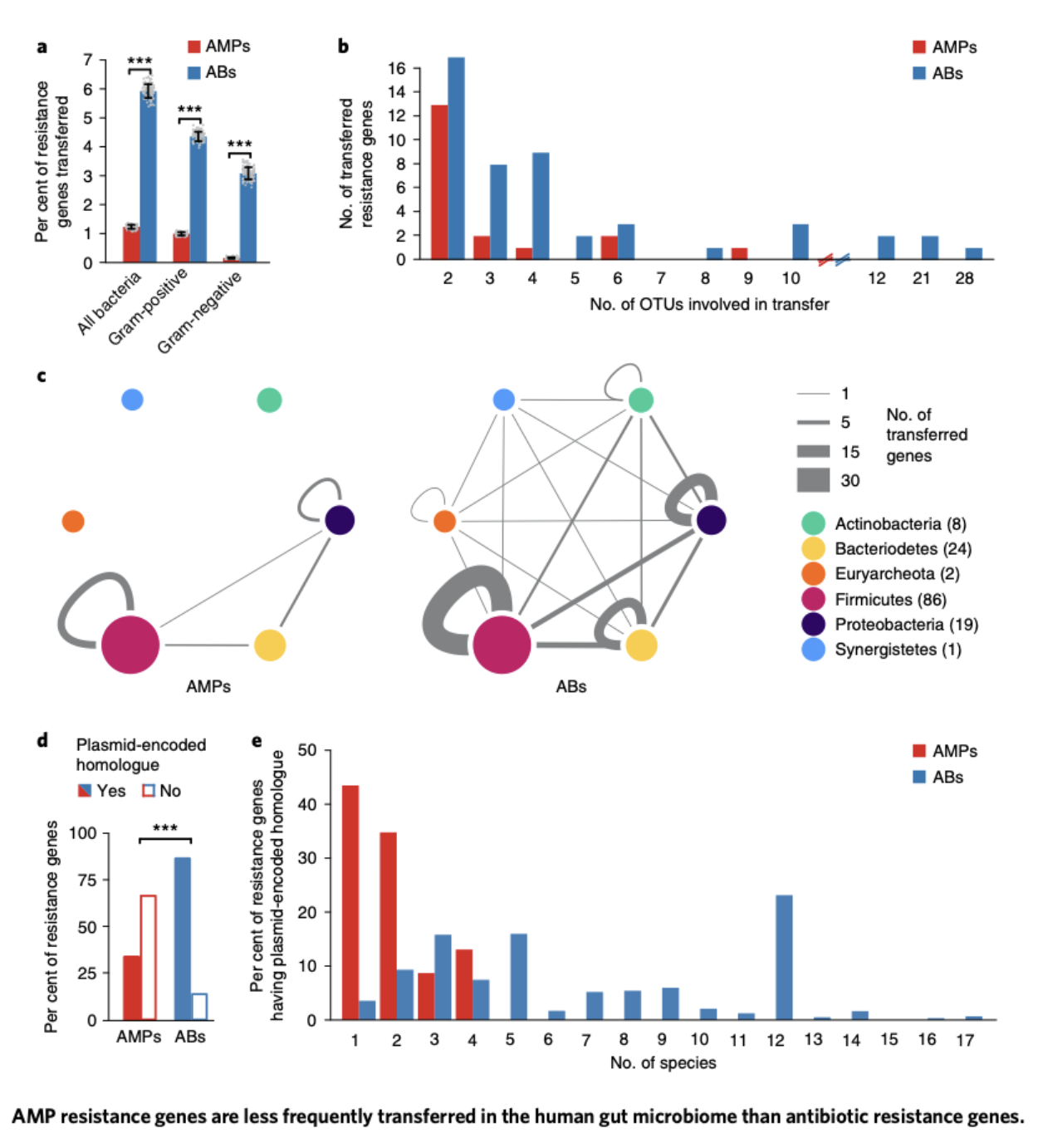
Bacteria can share antibiotic resistance genes through a process known as horizontal gene transfer. This mode of spreading mobile resistance genes has proven to be the primary mechanism by which bacteria protect themselves from the effects of antibiotics, significantly contributing to the ongoing antibiotic crisis.
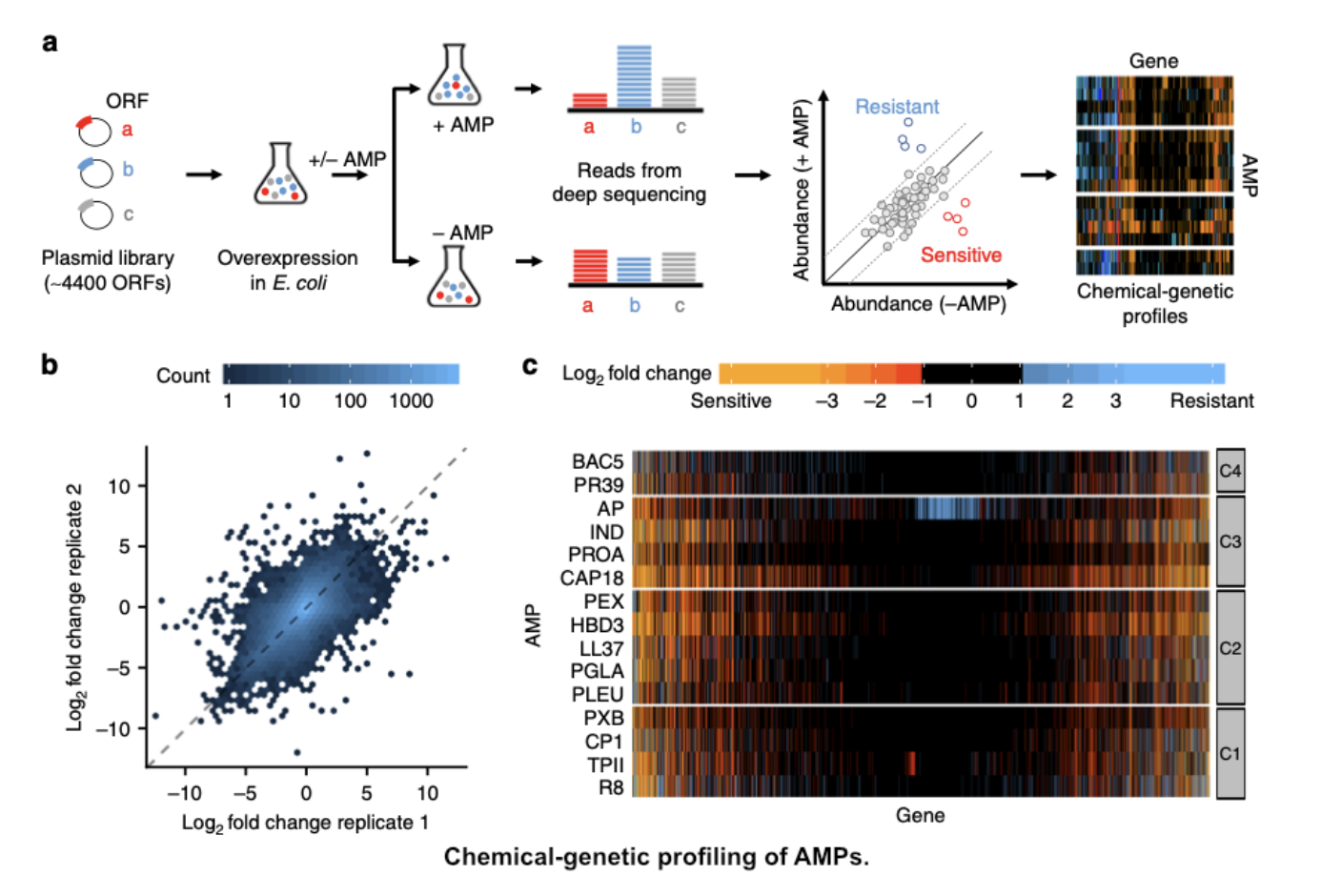
Our research focused on understanding the emergence of resistance genes against novel therapeutic approaches. We aimed to investigate how these resistance genes start to spread among human pathogens encountered in healthcare settings and within the human microbiota. One of our research projects, conducted in collaboration with the Pál and Papp Labs, centers on the horizontal transmission of resistance genes against cationic antimicrobial peptides (AMPs). While AMPs are promising therapeutic agents, they are also among the most ancient and efficient components of the innate immune defense strategy of multicellular organisms. This led us to the question why natural AMPs, integral to the human innate immune system, have remained effective for millions of years without the evolution of detectable resistance in several bacterial species. In contrast, clinical antibiotics tend to lose their effectiveness in a very short period of time.
Our study, published in Nature Microbiology, uncovered the presence of robust phylogenetic barriers to the horizontal transfer of AMP resistance genes in the human gut microbiota. This finding may provide a partial explanation for the persistence of AMP efficacy over extended evolutionary periods. Significantly, our work garnered recognition from the scientific community at Faculty 1000, and it has been included in the core collection of studies by the editorial board of Nature, signifying its importance in the context of gut microbiota research in 2019.
Augmenting Phage Therapeutic Potency by Shaping Phage-Human Interactions
Therapeutically applied phages often face challenges that compromise their effectiveness upon administration, greatly hindering their clinical utility. At the same time, our body is teeming with phages: apparently there are no human organs inaccessible to them, including those previously considered as ‘sterile’, such as the cerebrospinal fluid. There are clearly evolutionary pathways by which phages can infiltrate our body and thrive within. However, the specific mechanisms by which they accomplish this remain uncertain. One of our main research directions is to gain a deeper insight into human-phage interactions. This knowledge can be harnessed to greatly enhance the potency of phage therapies. Our research focuses on two clinically important aspects of phage activity in the human body. First, we investigate how phages can achieve elevated predatory potential and increased persistence in the gut. Second, we delve into how certain phages can easily bypass the otherwise infamously impenetrable Blood-Brain Barrier. Exploration and utilization of the underlying genetic factors enabled us to genetically modify clinically viable phages in a way that enhances their therapeutic potential. Beyond their evident value in eradicating bacterial diseases, the rational manipulation of phage-human interactions unlocks unanticipated possibilities that further expand the clinical applicability of phage-based therapeutics.
Balint Kintses established his laboratory in 2019, with support from the Hungarian Centre of Excellence for Molecular Medicine (HCEMM). With over 18 years of research experience, including a four-year postdoctoral tenure at the University of Cambridge, Kintses has extensive experience. He has more than 25 publications, mostly in high profile journals, such as Nature Microbiology and Nature Communications. Furthermore, he has played a pivotal role in the prototype development of several biochemical methodologies, such as advanced microfluidic-based high-throughput screening and genome engineering methodologies, many of which have achieved notable success on the global stage.

senior research associate

communication expert

PhD student

PhD student
PhD student

research fellow

research fellow

research fellow

PhD student

research assistant

PhD student

laboratory assistant

research assistant

PhD student

PhD student

MSc student

BSc student
 Bálint KINTSES
Bálint KINTSES
|
senior research associate | Publications | CV |
 Andrea FEJÉR
Andrea FEJÉR
|
communication expert | ||
 Tóbiás SÁRI
Tóbiás SÁRI
|
PhD student | ||
 Gábor APJOK
Gábor APJOK
|
PhD student | Publications | |
 Mihály KONCZ
Mihály KONCZ
|
PhD student | Publications | CV |
 Bálint VÁSÁRHELYI
Bálint VÁSÁRHELYI
|
research fellow | Publications | |
 Orsolya MÉHI
Orsolya MÉHI
|
research fellow | Publications | |
 Tamás STIRLING
Tamás STIRLING
|
research fellow | Publications | CV |
 Erika NAGRAND
Erika NAGRAND
|
PhD student | ||
 Dorina KOVÁCS
Dorina KOVÁCS
|
research assistant | ||
 András ASBÓTH
András ASBÓTH
|
PhD student | ||
 Izabella EGRI
Izabella EGRI
|
laboratory assistant | ||
 Sif Aldin ZALOKH
Sif Aldin ZALOKH
|
research assistant | ||
 Hiba HADJ Mehdi
Hiba HADJ Mehdi
|
PhD student | ||
 Dóra SALA
Dóra SALA
|
PhD student | ||
 Andrea LADÓ
Andrea LADÓ
|
MSc student | ||
 Kincső FELFÖLDI
Kincső FELFÖLDI
|
BSc student |