
Plants continuously form new organs during their whole life. This is due to the presence of the organ-forming meristematic regions. The main meristems are the apical meristems of the shoot and the root, which contribute to the continuous growth of the plant above- and underground. The meristems have also the function to lay down the lateral organs, the leaves, the flowers, and the side roots. These processes are dependent on the developmental program but are also strongly influenced by environmental signals. That is why there are no two plants looking identical despite having the same genetic material. The aim of our group is to better understand the flexible growth and developmental strategy of plants allowing them to adapt repeatedly to the changing environment. The goal is to identify some of the key molecular players that are central to control plant morphogenesis but are also capable of responding to environmental changes. We suppose that such molecular mechanisms allow the plants to adapt their form and function to the environmental conditions. In addition to increasing our basic biological knowledge, the research may open possibilities to develop improved plant varieties, which can either change or maintain their growth under unfavourable conditions depending on breeding aims. In our laboratory, among others, we investigate the developmental response of plants to increased ambient temperatures (thermomorphogenesis) in light of the forecasted climate change scenarios.
Transcriptional coordination of cell proliferations with quiescence and differentiation
The current view is that the entering and leaving the cell cycle are regulated by an evolutionarily conserved transcriptional mechanism called E2F-RB. According to the text book model, E2F transcription factors drive key cell cycle genes required for cell cycle entry, while the cell cycle inhibitory RETINOBLASTOMA tumor suppressor (RB) binds to E2Fs and stimulate cell cycle exit and establish quiescence. In Arabidopsis, a single RB-related protein (RBR) has been identified and that can form complexes with three E2F transcription factors (E2FA, E2FB, and E2FC). In the established model of plant cell cycle control, activator E2Fs are consistently portrayed as essential effectors of a transcriptional program that drives cells into cell cycle. We investigate the contribution of activator E2Fs to seed and embryo development in Arabidopsis. We find that, surprisingly, mutation of activator E2Fs (e2fab) has very little effect on the expression of cell cycle genes while seed storage proteins accumulate prematurely.

Figure 1. The temporal and spatial expression of WRINKLED 1 transcription factor (WRI1) in the developing embryo depend on its regulatory E2F-binding site. (A,B) Representative confocal images of developing embryos from the intact (A) and E2F-binding site mutant (B) WRI1 promoter (pWRI1::CFP and pmutE2FWRI1::CFP reporter lines, respectively) dissected from immature seeds.
Thus, E2Fs coordinate the switch between the morphogenic and the maturation phase of seed development by preventing the untimely expression of genes specific to each phase. By studying the function of E2FB during leaf development we find that E2FB regulates its up-stream regulators RBR and CYCLIND3;1. These autoregulatory loops should stabilize the balance between proliferation and differentiation and prevent the development of tumorous growth. Plant RBR together with E2FA are involved in the maintenance of genome stability, while E2FB and E2FC form complexes together with RBR and DREAM components (Fig.2). In various animals related multiprotein DREAM complexes were identified to be in control of cell cycle.
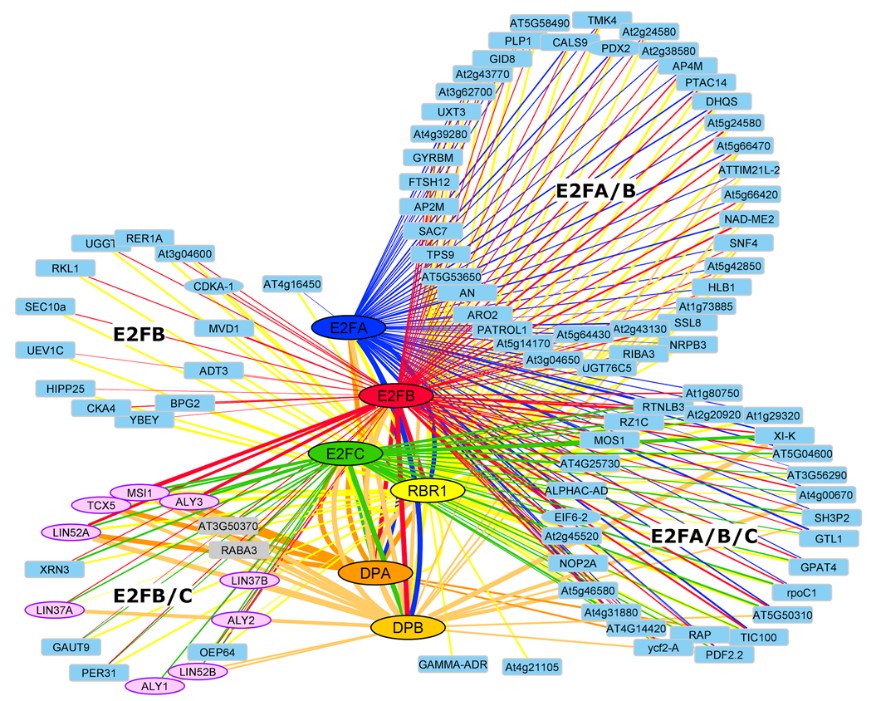
Figure 2. Meta-Analysis of FP-IP results from seedlings.
Cytoscape representation of a meta-analysis of 182 FP-IPs using different bait proteins (35 × E2FA-GFP [blue], 39 × E2FB-GFP [red], 20 × E2FC-GFP [green], 32 × RBR1-GFP [yellow], 3 × DPA-GFP [orange], and 3 × DPB-CFP [pale-orange], 50 × GFP [control]). To reduce complexity, only prey proteins which were enriched in at least one third of the IPs of one bait are shown. The thickness of the edges corresponds to the frequency of positive IPs by which a bait/prey interaction was found. Proteins that were also identified in the tandem affinity purification experiments are represented by an ellipse. Prey proteins are grouped according to their occurences in different E2F-IPs. DREAM complex homologs are shown in dark blue. Additional proteins that display an interaction pattern like the DREAM component homologs, that is, do not interact with E2FA, but interact with E2FB/C and the DPs, are shown in bold with a thick outline.
In plants, there is a tight balance among cells proliferating in meristematic regions of emerging young organs, stem cells that are in a reversible quiescent status with a capacity to re-entry into cell division upon stimulation, and the most common cellular state of terminally differentiated cells formed after permanent exit from the cell cycle. The principal regulation of cell proliferation by highly conserved molecular mechanisms is well established, but how the cell cycle machinery is controlled in quiescent cells to maintain transient and stable repression of mitosis remains unclear. By studying triple e2fabc mutant Arabidopsis plants, surprisingly we observed hyperplasia just as upon RBR silencing suggesting that E2Fs have repressive role. RBR silencing leads to growth arrest during seedling establishment due to the inhibition of this developmental transition and of cellular differentiation. This is not the case for the e2fabc mutant, which shows hyperplasia during embryonic and post-embryonic organ development with delayed cell cycle exit and unstable quiescence both in stem and in terminally differentiated cells (Fig.3).
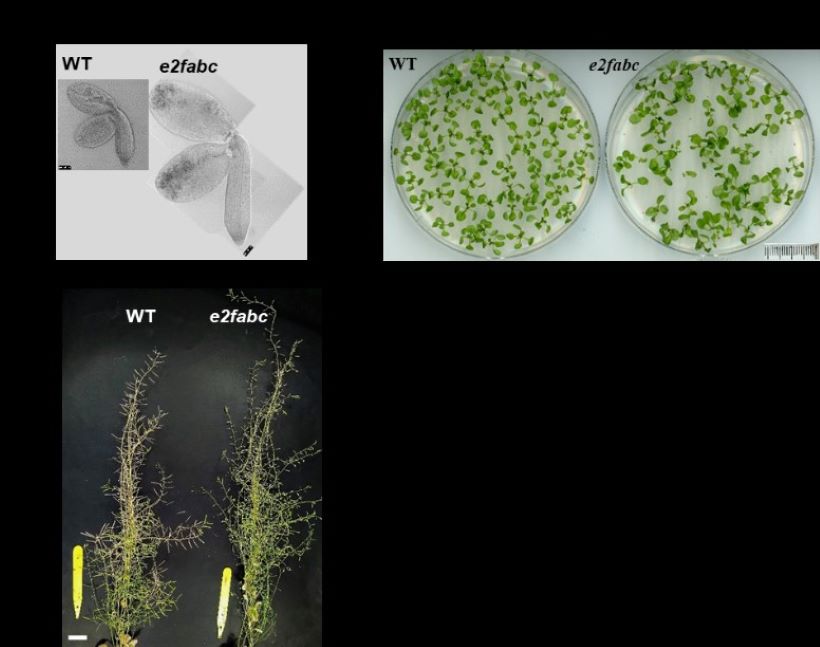 |
Figure 3. The e2fabc mutant plants grow taller and develop larger organs. A, E2fabc embryos are larger than those of WT. B, Representative phenotypes of plantlets at 11 DAG grown on agar medium, or C, mature plant grown on soil showing that the e2fabc mutant has shorter siliques and is taller than the WT.
Accordingly, puzzle form differentiated pavement cells in the epidermis of e2fabc leaf were found in division indicating that terminal quiescence was neither properly established nor maintained in the absence of E2Fs. Additionally, the rarely dividing quiescent centre cells in the stem cell niche region of root meristem became more proliferative in the triple e2fabc mutant root than in the WT supporting that transient quiescence was unstable (Fig.4).
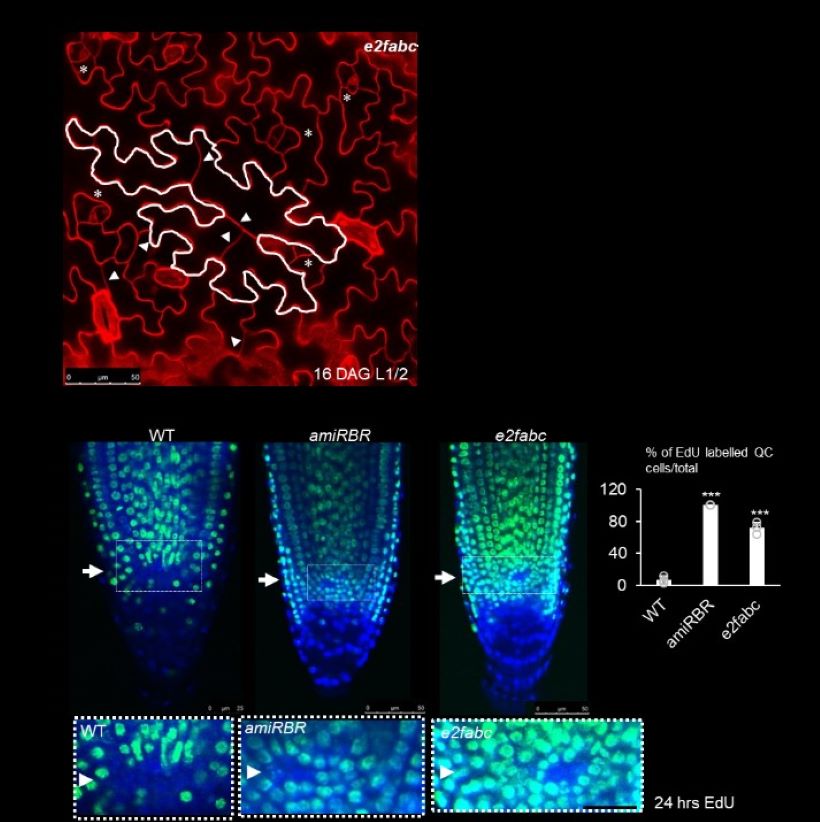
Figure 4. Quiescence is regulated by E2Fs.
A, The e2fabc mutant leaves display extra cell division events in puzzle-shaped differentiated epidermal cells. An example of a large dividing puzzle-formed pavement cell is outlined in white. Arrowheads show straight cell lines indicative for multiple extra cell divisions. Asterisks mark dividing small meristemoid-like cells. Picture of leaf epidermis was taken at 16DAG. B, Cell proliferation is reactivated in the QC of e2fabc and amiRBR lines. Plants were incubated in EdU for 24h. Representative images are shown on the top panel and the white arrows point at the QC. White boxes outline the QC region of root meristems and magnified in below. White arrowheads mark the QC. The graph below shows the proportion of roots showing EdU labelling in the QC (n = 100 samples in each).
We have also shown that the canonical E2Fs together with RBR form a repressor complex on a cohort of cell cycle genes that is pivotal for maintaining quiescence. Through this repression mechanism it is possible to influence plant growth which could open novel ways for improving crop biomass and productivity.
Signaling pathways affecting plant morphogenesis
Plants exhibit a remarkable developmental plasticity as a consequence of their sessile way of life. During plant ontogenesis, exogenous (environmental) and endogenous (developmental) signals converge on interlinked signaling pathways having dynamic impacts on plant form and function. We use molecular, biochemical and cellular approaches to reveal and characterize plant-specific pathways of cellular signaling that underlie this developmental plasticity. We aim to reveal the link of ROP GTPases to various kinases (upstream/downstream) in order to have new insights into the regulation of cell growth, polarity, and differentiation during plant morphogenesis. We provided the first experimental data that ROPs can regulate plant-specific receptor-like cytoplasmic kinases (RLCKs) and they might also be regulated through phosphorylation. We aim to use the obtained knowledge to build up a general model for receptor signaling in plants; from receptor kinases to ROP GTPase effectors.
In vitro plant regeneration
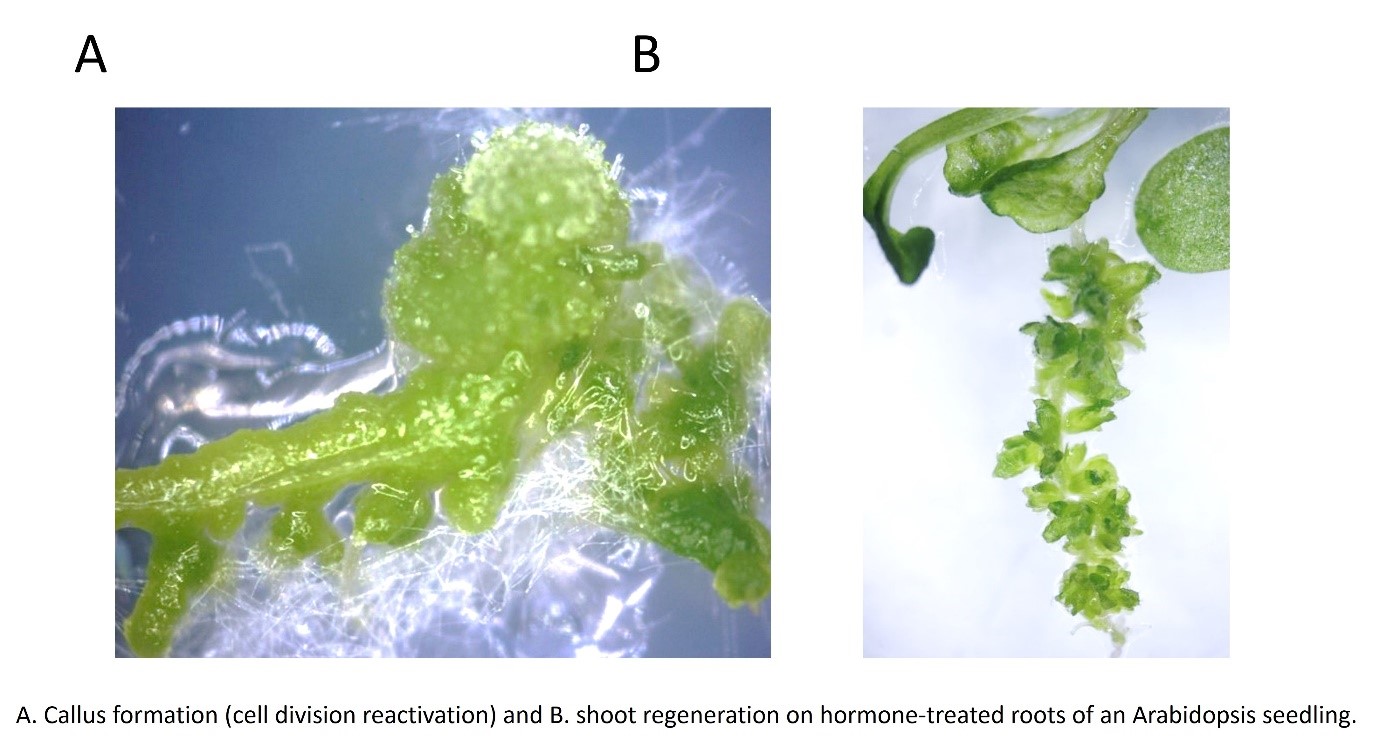
One of the most intriguing examples of plants developmental plasticity is in vitro plant regeneration either via somatic embryogenesis or organogenesis during which differentiated plant cells develop into embryos, shoots, or roots, respectively. Despite the fact that these processes are widely used for in vitro plant propagation, the biological background of these plant-specific phenomena is hardly known.
Although plant cell and tissue culture has already a long history and is widely used in biotechnology, our knowledge about plant cell regeneration is scarce. We still do not know how differentiated somatic cells lose differentiated functions and regain the ability to divide (i.e. dedifferentiate) during culture. Our previous results indicate that this process might be governed by the same factors that regulate the cell division/differentiaon switch during in planta development. Using mutants and transgenic lines we will support this hypothesis first in Arabidopsis. ). In cereals, adult tissues are recalcitrant to dedifferentiate preventing their use in biotechnology. Based on the information obtained with Arabidopsis, we intend to determine the nature of the dedifferentiation block and to provide solution to overcome it using transient gene expression in these cells. In addition, we study the role of hormonal, molecular, polyamine-dependent, and redox regulation of in vitro plant regeneration.
Root development in Brachypodium, role of LOB-domain transcription factors
In this project, we use mainly Brachypodium distachyon, the model system of grasses including important cereal species: wheat, barley and rye. Our aim is to understand how the fundamental decision of cells to "divide or differentiate" is regulated in plants. We have chosen root development as the developmental process to study, as the development of a strong and efficient root system is crucial for plant survival under water scarce environmental conditions.
A wide range of Brachypodium genotypes have been tested for their drought tolerance and the selected lines are characterized in detail by determining their root growth and architecture in response to water deficit and temperature changes. We have shown that water deficit alters the circadian clock function in the root system and that root proline metabolism is regulated differently from that of the shoot during water limitation.
Among the candidate genes that influence root architecture, we are mainly studying the so-called LOB (lateral organ boundary) domain transcription factor (TF) gene family of purple false brome (B. distachyon), which consists of 28 members (LBD genes) in this species, and their basic expression characterization has been performed in recent years. We screened for LBD genes that exhibit altered expression in segments of the root during thermo-morphogenic changes. We are currently investigating the interactions of LBD proteins mainly by computer modelling, but we also want to explore their possible phosphorylation by the cyclin/cdk complex, which requires the study of CDK/cyclin and E2F/RB regulatory pathways in Brachypodium too. We plan to characterize LBD mutants phenotypically and to analyze the expression of cell cycle regulatory genes and LOB domain transcription factor genes in these mutants.
The expected results may contribute significantly to our knowledge in the field of plant developmental biology. Despite the fact that Arabidopsis developmental biology has shown tremendous advances as a dicotyledonous plant, many questions remain unanswered with this excellent model plant. In addition, it is well known, partly thanks to our previous results, that there are substantial differences in cell cycle regulation between monocots and dicots. This fact also necessitates the use of a monocot model, so that we can compare the results with those obtained on Arabidopsis. Understanding the molecular genetic background of root development is also of high importance from a practical point of view. Brachypodium, as a closely related model plant for wheat, barley, and rye, allows us to apply the knowledge we have acquired on this small, fast-growing, and simple grass to the above crops of great agronomic importance.

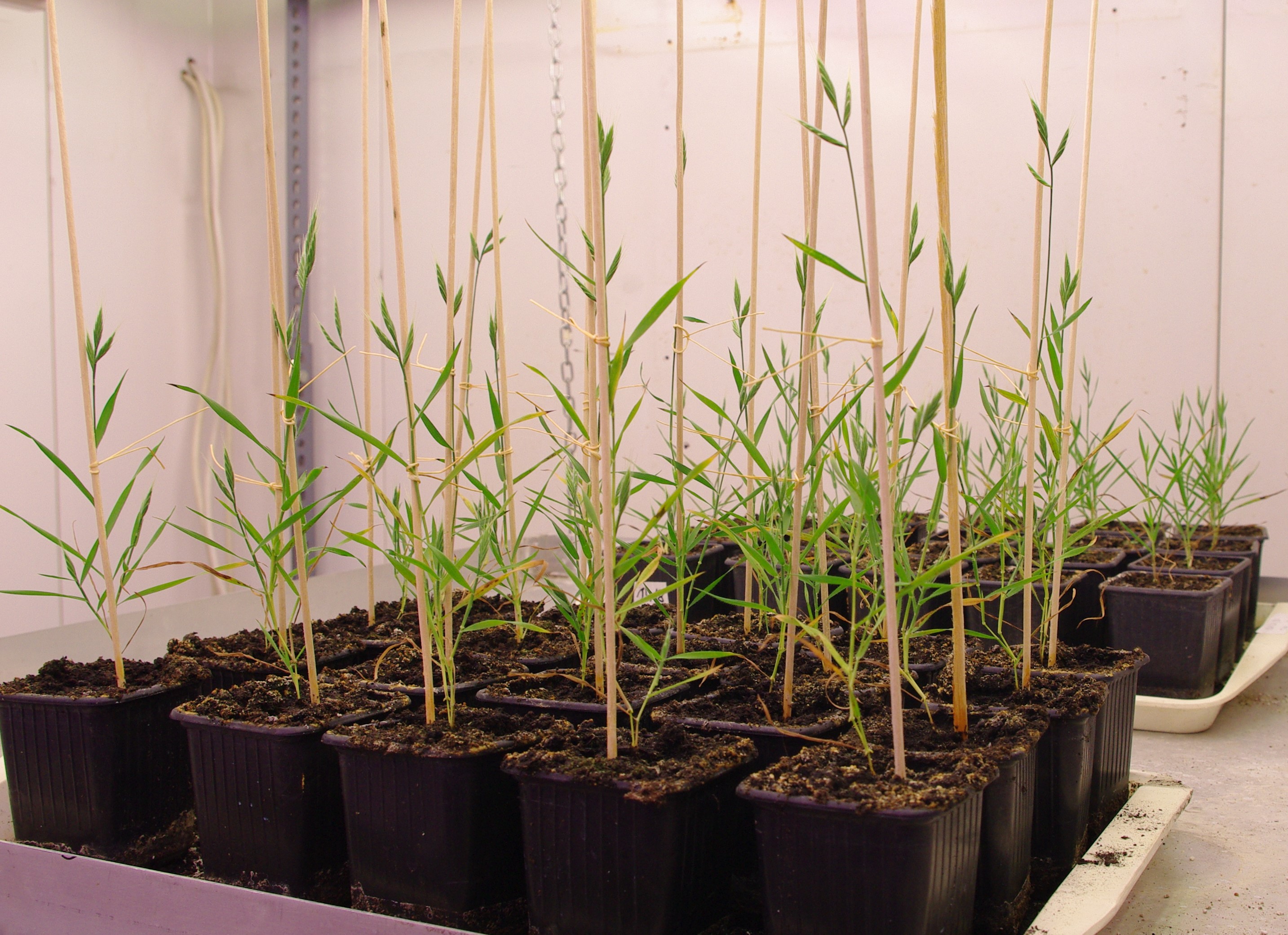
Purple false brome plants (Brachypodium distachyon) at vegetative and flowering stage, grown in plant growth chamber.

senior research associate

scientific adviser

senior research associate

research fellow

research fellow

reserach fellow

junior research associate

junior research associate

PhD student

guest researcher

technical assistant

laboratory assistant
 Zoltán MAGYAR
Zoltán MAGYAR
|
senior research associate | publications | CV |
 Attila FEHÉR
Attila FEHÉR
|
scientific adviser | publications | CV |
 János GYÖRGYEY
János GYÖRGYEY
|
senior research associate | publications | CV |
 Erzsébet KENESI
Erzsébet KENESI
|
research fellow | publications | CV |
 Eszter MOLNÁR
Eszter MOLNÁR
|
research fellow | publications | CV |
 Magdolna GOMBOS
Magdolna GOMBOS
|
reserach fellow | publications | CV |
 Péter BENKŐ
Péter BENKŐ
|
junior research associate | publications | CV |
 Fruzsina NAGY
Fruzsina NAGY
|
junior research associate | publications | CV |
 Attila HLACS
Attila HLACS
|
PhD student | CV | |
 Katalin GÉMES
Katalin GÉMES
|
guest researcher | publications | |
 Edina KISS
Edina KISS
|
technical assistant | ||
 Róza NAGY
Róza NAGY
|
laboratory assistant |