
Regarding mortality, heart and blood vessel diseases are followed by cancer. Hungary has the highest rate of cancer-related mortality in the European Union; cancer is responsible for over 26% of deaths now. Although it is common to classify the tumours by their tissue of origin or their metastatic properties, all tumours share the characteristic feature of originating from one of our own cells.

To make the transition from normal to cancerous, cells have to change some of their basic functions. We know that increased exposure to mutagens increases the risk of cancer, and that mainly the DNA-damaging properties of the mutagens are responsible for this. Altered information in the genome leads to altered proteins. These altered proteins can gain new functions that can release the cells from their normal cell cycle and push them to uncontrolled continuous proliferation. Then, the cells that are able to ensure their nutritional supply and survive in different tissue environments are selected by selective pressure.
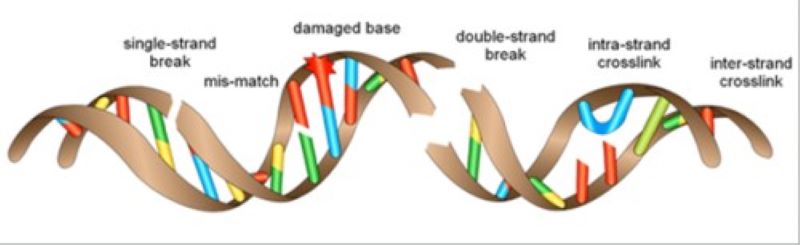
Covalent modifications of DNA by carcinogens do not always lead to mutations; several hundred thousand of modified bases arise in a cell every day. DNA repair mechanisms prevent the conversion of DNA damage to mutations.
Our research group studies the processes that represent the last chance for the cell to survive the devastating effects of unrepaired base modifications. Covalently modified bases pose a barrier for the replication machinery during DNA replication, which it can only overcome by inserting a non-fitting nucleotide. Such events occur after UV irradiation in skin cells when UV irradiation forms covalent cross-links between neighbouring bases, e.g., thymines or cytosines. The polymerase responsible for the replication of DNA is unable to read these modified bases correctly and to insert correct nucleotides opposite them. Stalling of the replication fork may result in cell death; to prevent it, the cell can activate the pathway governed by the Rad6/Rad18 enzyme complex, which results in the replacement of the replicative polymerase by a TLS polymerase such as Polη. Translesion polymerases are capable of resuming DNA synthesis by inserting the correct adenines opposite the cross-linked thymines. However, in many cases these TLS polymerases insert an incorrect nucleotide opposite DNA lesions. In every such case, the price of resuming replication and the survival of the cell is picking up a new mutation. A mutation can be a fair price for survival provided the information stored in the position concerned is not essential for the daughter cells.
There is a mechanism capable of rescuing the stalled replication fork in an error-free manner, but the process involves vulnerable DNA structures; therefore, it is not always suitable. Our group was the first to show that the Rad5 enzyme is capable of catalysing such reactions (1). Also, we were among the first groups to prove that the HLTF and SHPRH genes are the human homologues of the yeast Rad5 (2, 3, 4). In human cells, the HLTF protein is inactive in many tumour types, and its absence increases the frequency of chromosome aberrations, indicating its significance in genome stability and tumour suppression. Our group demonstrated the in vivo role of the HLTF protein in the process of postreplication repair and characterized the roles of its different domains (5).
The Spartan protein plays a regulatory role in the DNA damage tolerance pathway; however, its exact function was unknown. Our group demonstrated that Spartan participates in the replication of DNA-protein crosslink-containing DNA: due to its DNA-dependent protease activity, it removes crosslinked proteins. We also revealed a new biochemical characteristics of the Spartan protein by demonstrating the DNA-binding ability of the purified human protein and proving its function in the recruitment of the Polη translesion polymerase to the site of DNA damage (6).
These newly identified proteins are perfect targets for tumour therapeutic agents since most of the currently available chemotherapeutics (cysplatin, mitomycin C, etc.) act by damaging DNA and blocking the replication in rapidly dividing cancer cells. It is conceivable that by inhibiting the mechanisms that resolve stalled replication forks more cancer cells can be eliminated.
The regulation of the processes taking place upon replication fork stalling is executed by a highly complicated, oversecured, and sensitive mechanism since changing the correct genomic information should only be the last option. For example, our research group has evidence from in vivo localization and immune-precipitation experiments and also from in vitro protein-protein interaction experiments for the participation of protein factors in the Rad6-Rad18 pathway that have not been identified yet (7, 8) or have been identified in connection with other pathways and diseases like Fanconi anaemia.
In our laboratory, we have developed a new method with which we are able to follow the kinetics of the resolution of the stalled replication forks. This enables us to examine the regulatory role of a given protein in knockdown or overexpression experiments and to reveal the process that can replace the functions that were knocked out.
The missing or erroneous activity of protein factors involved in the replication of damaged DNA directly leads to a higher probability of cancer. In some cases, cancer shows familiar occurrence; however, its cause often remains unknown. Next-generation sequencing (NGS) techniques provide information regarding the origins of the high probability of cancer. With this technique in our hands, we can obtain sequence data from several genes in a high number of patients simultaneously. Furthermore, we can map mutations of different genes in different parts of the tumour, along with the mutation rate.

Since these genes are directly involved in the development of mutations, it is interesting to see how their overexpression or knockout influences the mutation rate in cells. We are currently examining the impact of different mutagens or the elimination of protein factors on the frequency and quality of mutations occurring in reporter genes. A simple answer to the well-known question: “When will the cure for cancer be found?” does not exist since each tumour has to be treated as a different entity, and the genetic background in a tumour can differ even from cell to cell. However, a complex answer can be given; with time, we will identify newer and newer targets; therefore, in the end, personalized medicine and therapy combined with early identification will lead to the harmless outcome of a currently deadly disease.
Selected publications:
1. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Mol Cell. 2007 Oct 12;28(1):167-75.
2. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Unk I, Hajdú I, Fátyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3768-73. doi: 10.1073/pnas.0800563105. Epub 2008 Mar 3.
3. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Blastyák A, Hajdú I, Unk I, Haracska L. Mol Cell Biol. 2010 Feb;30(3):684-93. doi:10.1128/MCB.00863-09. Epub 2009 Nov 30.
4. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Achar YJ, Balogh D, Haracska L. Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14073-8. doi:10.1073/pnas.1101951108. Epub 2011 Jul 27.
5. Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Achar YJ, Balogh D, Neculai D, Juhasz S, Morocz M, Gali H, Dhe-Paganon S, Venclovas Č, Haracska L. Nucleic Acids Res. 2015 Dec 2;43(21):10277-91. doi: 10.1093/nar/gkv896. Epub 2015 Sep 8.
6. DNA-dependent protease activity of human Spartan facilitates replication of DNA-protein crosslink-containing DNA. Mórocz M, Zsigmond E, Tóth R, Enyedi MZ, Pintér L, Haracska L. Nucleic Acids Res. 2017 Apr 7;45(6):3172-3188. doi: 10.1093/nar/gkw1315. PMID: 28053116
7. Role of SUMO modification of human PCNA at stalled replication fork. Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, Szukacsov V, Burkovics P, Haracska L. Nucleic Acids Res. 2012 Jul;40(13):6049-59. doi: 10.1093/nar/gks256. Epub 2012 Mar 28.
8. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Nucleic Acids Res. 2012 Nov;40(21):10795-808. doi: 10.1093/nar/gks850. Epub 2012 Sep 16.
9. The Combination of Single-Cell and Next-Generation Sequencing Can Reveal Mosaicism for BRCA2 Mutations and the Fine Molecular Details of Tumorigenesis. Gráf A, Enyedi MZ, Pintér L, Kriston-Pál É, Jaksa G, Bálind Á, Ezer É, Horváth P, Sükösd F, Kiss E, Haracska L. Cancers (Basel). 2021 May 13;13(10):2354. doi: 10.3390/cancers13102354. PMID:34068254
10. Coordinated Cut and Bypass: Replication of Interstrand Crosslink-Containing DNA. Li Q, Dudás K, Tick G, Haracska L.Front Cell Dev Biol. 2021 Jun 28;9:699966. doi: 10.3389/fcell.2021.699966. eCollection 2021.PMID: 34262911
11. BC-Monitor: Towards a Routinely Accessible Circulating Tumor DNA-Based Tool for Real-Time Monitoring Breast Cancer Progression and Treatment Effectiveness. Priskin K, Pólya S, Pintér L, Jaksa G, Csányi B, Enyedi MZ, Sági-Zsigmond E, Sükösd F, Oláh-Németh O, Kelemen G, Nikolényi A, Uhercsák G, Sántha D, Dobi Á, Szilágyi É, Valicsek E, Tordai L, Tóth R, Kahán Z, Haracska L.Cancers (Basel). 2021 Jul 12;13(14):3489. doi: 10.3390/cancers13143489.PMID: 34298704
12. A series of xanthenes inhibiting Rad6 function and Rad6-Rad18 interaction in the PCNA ubiquitination cascade.Fenteany G, Sharma G, Gaur P, Borics A, Wéber E, Kiss E, Haracska L.iScience. 2022 Mar 10;25(4):104053. doi: 10.1016/j.isci.2022.104053. eCollection 2022 Apr 15.PMID: 35355521
13. A Comprehensive Evaluation of the Performance of Prediction Algorithms on Clinically Relevant Missense Variants.Qorri E, Takács B, Gráf A, Enyedi MZ, Pintér L, Kiss E, Haracska L.Int J Mol Sci. 2022 Jul 19;23(14):7946. doi: 10.3390/ijms23147946.PMID: 35887294

scientific adviser

senior research associate

research associate

research associate

laboratory administrator

PhD student

PhD student

PhD student

PhD student

PhD student

administrator expert

administrator expert

researcher

technician
 Lajos, HARACSKA
Lajos, HARACSKA
|
scientific adviser | publications | CV |
 Ernő, KISS
Ernő, KISS
|
senior research associate | publications | CV |
 Mónika, MÓROCZ
Mónika, MÓROCZ
|
research associate | publications | CV |
 Alexandra, GRÁF
Alexandra, GRÁF
|
research associate | publications | CV |
 Gabriella, ÁDÁMNÉ TICK
Gabriella, ÁDÁMNÉ TICK
|
laboratory administrator | publications | |
 Bertalan Vilmos, TAKÁCS
Bertalan Vilmos, TAKÁCS
|
PhD student | publications | CV |
 Emese, PEKKER
Emese, PEKKER
|
PhD student | publications | CV |
 Erda, QORRI
Erda, QORRI
|
PhD student | publications | CV |
 Ádám Tamás, SÁNTA
Ádám Tamás, SÁNTA
|
PhD student | publications | CV |
 Valentin, VARGA
Valentin, VARGA
|
PhD student | publications | CV |
 Katalin, KOVÁCS
Katalin, KOVÁCS
|
administrator expert | ||
 Katalin, VINCZE-KONTÁR
Katalin, VINCZE-KONTÁR
|
administrator expert | publications | CV |
 Katalin PRISKIN
Katalin PRISKIN
|
researcher | ||
 Erika LACZ
Erika LACZ
|
technician |