
Introduction of the Laboratory of Bacterial Physiology and Strain Engineering
The turn of the millennium brought the advent of whole genome sequencing, initially for microbial cells, and later for cells of higher-order organisms. Next generation sequencing further eased the financial and technical limits of sequence-acquisition. The consequential acceleration in the growth of genomic sequence data introduced a revolution in at least three scientific subdivisions: i) evolutionary biology, where sequence data warranted the construction of phylogenetic trees of previously unseen detail, ii) systems biology, where the building of genome-scale models finally became a reality, and iii) molecular genetics, where the exact and complete DNA sequences permitted the targeted modification of any gene of choice. Our workgroup concentrates on the third subdivision, but nevertheless interacts with other laboratories of the Unit to support or utilize findings of evolutionary and systems biology as well. In brief, we participate in the development of experimental genomics tools, and apply it for two complementary tasks: revealing information on the genetic organization of bacterial cells, and using it to develop, optimize and maintain bacterial cell factories for biotechnological purposes.
In detail:
Our projects currently fall into two major categories. The first one investigates mobile genetic elements, while the second one concentrates on microbial biotechnology.
Transposable elements: parasites or saviors?
One of the most surprising discoveries of the 20th century was the observation that certain genes are capable of transposition, that is jumping from one position to the other on the chromosome. Such mobile elements, also called transposable elements are investigated from multiple aspects. A long-standing question of evolutionary biology, is whether the invasion of host populations by these elements resulted from the selective advantage they conferred to their hosts, or was mereley a result of their selfish, replicative nature. The importance of transposable elements from the aspect of human health is paramount. As a top concern, they assist transferring certain genes from one bacterial strain to the other thereby providing protection against antibiotic treatment. Therefore, understanding and controlling their transposition may introduce a novel strategy to fight the worldwide emergence of pathogenic bacteria resistant to multiple antibiotics.
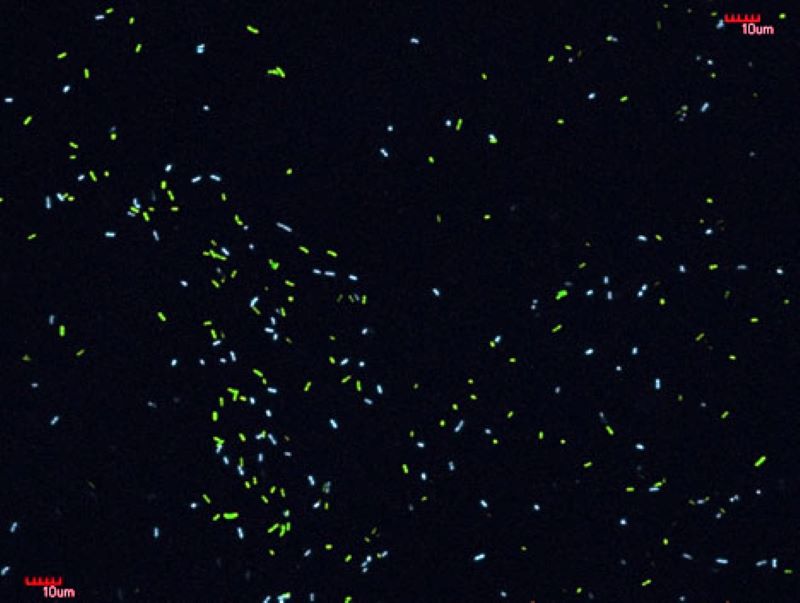
Fluorescent microscopic image of a mixture of E. coli cells expressing YFP or CFP proteins.(Fehér et al. 2012 Mol Biol Evol)
Metabolic engineering: genetic reprogramming of metabolic networks
Metabolic engineering is often defined as the reprogramming of cells to produce, break down or transform compounds of interest. This scientific field emerged in the 1990s, and is today supporting a worldwide industry. The goal of our young team is to develop and test novel genetic methods to expand the toolbox of metabolic engineering. We are interested in multiple divisions of the engineering workflow: modification of endogenous genes, cloning and establishment of exogenous pathways, gene expression control, diversity generation, screening and selection, modulation of genetic plasticity, etc... We rely on earlier experience gathered in genome editing during the engineering of a multi-deletion E. coli strain, and during the verification of an in silico tool developed for metabolic pathway design. Our aim is to demonstrate the utility and efficiency of the extended toolbox through the producion of novel added-value chemicals.
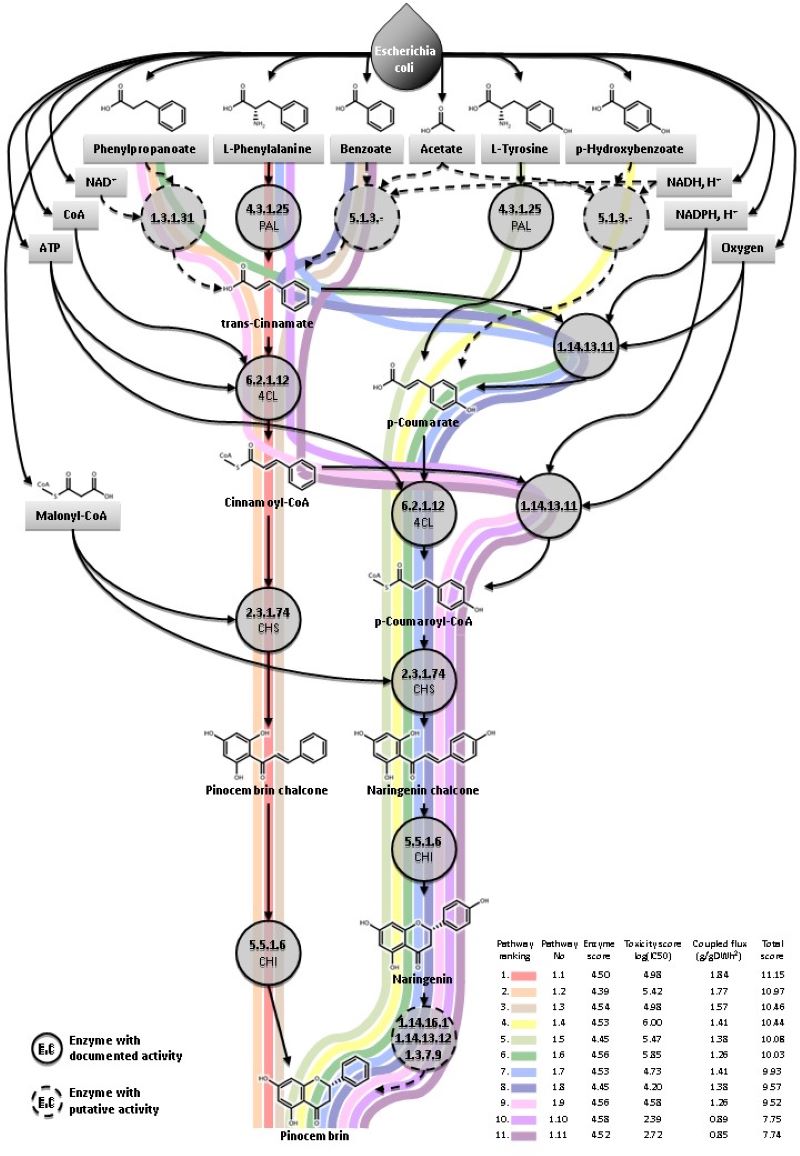
Pathways predicted to produce the plant flavonoid Pinocembrin in E. coli. (Fehér et al. 2014 Biotech J.)
Transposon-silencing
Stabilizing bacterial genomes has been our passion for nearly two decades. We have participated in projects that have eliminated mobile genetic elements from the genome of Escherichia coli K-12 MG1655 (Pósfai et al., 2006), and helped to demonstrate the longer half-life of toxic protein-encoding genes within this strain (Umenhoffer et al, 2010). We have also contributed to the project that transferred the clean genomic regions of E. coli K-12 to BL21 to generate a protein-production host free of mobile genetic elements (Umenhoffer et al, 2017). Another similar endeavor we were connected to eliminated error-prone DNA polymerases to reduce the rate of point mutations (Csörgő et al., 2012). But all these projects consumed a significant amount of time and effort to obtain increased genetic stability in bacteria. In this current project, we display the near complete silencing of the transposition of four bacterial insertion sequences: IS1, IS3, IS5 and IS150. We use the inactivated Cas9 protein (dCas9) to specifically bind the left inverted repeat of these IS elements in vivo. We demonstrate the decreased transcription rate of three of the four transposase types, but nevertheless reduce the transposition frequency of all four IS types to nearly zero. The portability of this technique is showcased by the efficient downregulation of transposition in multiple E. coli strains. Besides increased genomic stability, these strains also display a longer half-life for the production of certain toxic heterologous proteins. To the best of our knowledge, this is the first portable system applicable for the silencing of four different mobile genetic elements in parallel. For more details, please see our recent publication (Nyerges et al., 2019)!
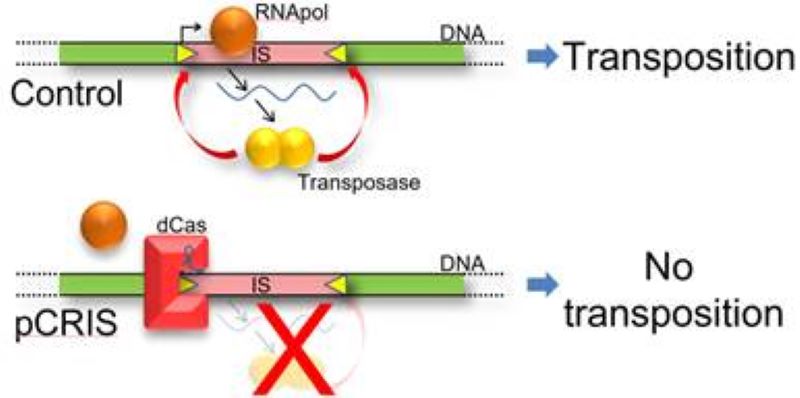
The mechanism of transposon silencing using CRISPR/dCas (Nyerges et al., 20`18 Synth Biol)
Uncovering a case of heritable progressive myopia
As an outlook to further scientific fields, we are also involved in a collaborative human genetic study, initiated by Dr. Noémi Széll. We investigated the unusual heredity pattern of an early onset high myopia disease, which manifests only in female members of the affected family. Based on the pedigree, we identified the carrier males and observed that all female offspring of the carrier males are consistently affected. Affected females, on the other hand had both affected and healthy daughters. This allowed us to hypothesize that the pattern of heredity is X-linked dominant, female limited. Next, Seqomics Biotechnology Ltd. carried out whole exome sequencing on two individuals and the Clinical Genetics Laboratory of the University of Szeged analyzed the raw sequences to identify a nonsense mutation in the ARR3 gene, located on the X-chromosome. This is the first description of a Caucasian family displaying ARR3-linked early onset high myopia. We are currently in the process of uncovering the molecular mechanisms leading to ocular elongation and a consequential myopia in our patients.
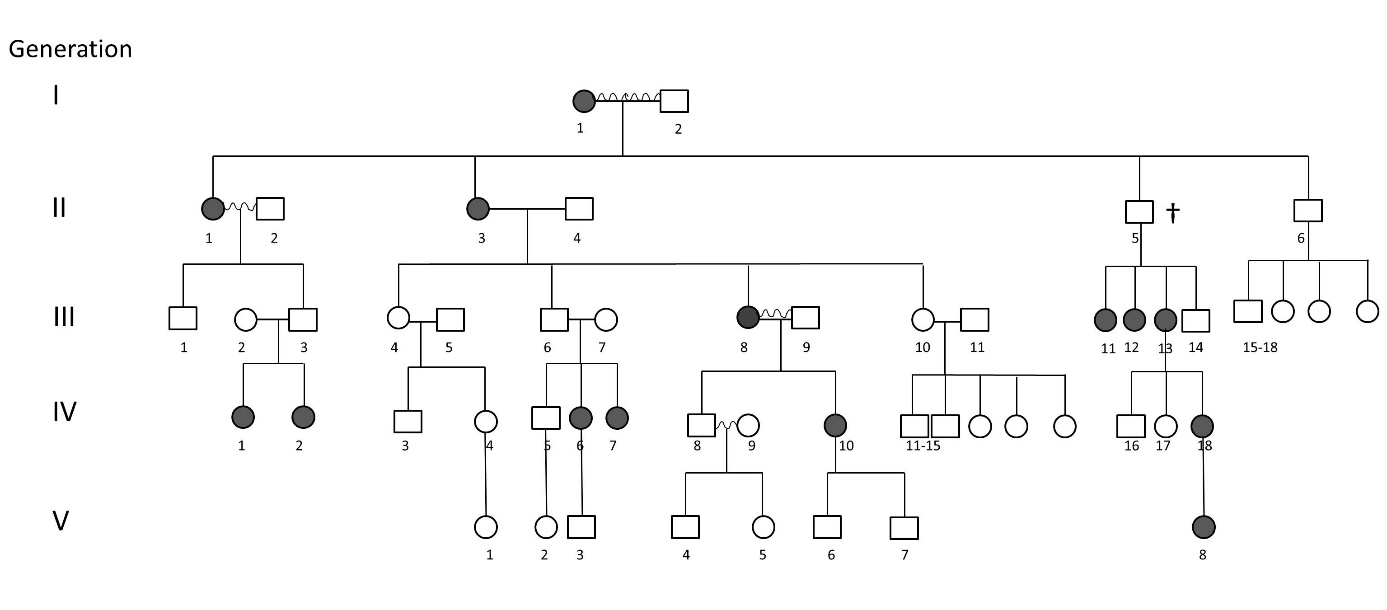
Pedigree of the family displaying female-limited early onset high myopia.

Senior Research Associate

Senior Research Associate

PhD student

PhD student

PhD student
 Tamás FEHÉR
Tamás FEHÉR
|
Senior Research Associate | publications | CV |
 Ferenc ÖTVÖS
Ferenc ÖTVÖS
|
Senior Research Associate | publications | CV |
 Ákos AVRAMUCZ
Ákos AVRAMUCZ
|
PhD student | publications | CV |
 Zsuzsanna Katalin VARGA
Zsuzsanna Katalin VARGA
|
PhD student | CV | |
 Dávid GOMBOS
Dávid GOMBOS
|
PhD student | publications |